Gene-wise codon usage and bias analysis of SARS-CoV-2 (2019-nCoV)
| No |
codon |
AA |
ORF1ab |
ORF1a |
S |
E |
M |
N |
ORF3a |
ORF6 |
ORF7a |
ORF7b |
ORF8 |
ORF10 |
| 1 |
TTT |
F |
254 |
156 |
59 |
2 |
5 |
3 |
8 |
3 |
7 |
3 |
4 |
3 |
| 2 |
TTC |
F |
95 |
52 |
18 |
3 |
6 |
10 |
6 |
0 |
3 |
3 |
4 |
1 |
| 3 |
TTA |
L |
201 |
117 |
28 |
1 |
4 |
2 |
3 |
3 |
2 |
3 |
6 |
0 |
| 4 |
TTG |
L |
116 |
87 |
20 |
2 |
4 |
9 |
9 |
0 |
1 |
1 |
2 |
1 |
| 5 |
CTT |
L |
185 |
129 |
36 |
7 |
12 |
8 |
10 |
1 |
6 |
4 |
2 |
0 |
| 6 |
CTC |
L |
61 |
39 |
12 |
0 |
6 |
2 |
5 |
2 |
2 |
0 |
0 |
2 |
| 7 |
CTA |
L |
77 |
45 |
9 |
2 |
5 |
3 |
1 |
2 |
1 |
1 |
0 |
1 |
| 8 |
CTG |
L |
28 |
18 |
3 |
2 |
4 |
3 |
2 |
0 |
3 |
2 |
0 |
0 |
| 9 |
ATT |
I |
169 |
109 |
44 |
1 |
11 |
9 |
9 |
5 |
4 |
4 |
5 |
0 |
| 10 |
ATC |
I |
57 |
38 |
14 |
1 |
6 |
4 |
5 |
1 |
1 |
1 |
5 |
0 |
| 11 |
ATA |
I |
117 |
68 |
18 |
1 |
3 |
1 |
7 |
4 |
3 |
0 |
0 |
3 |
| 12 |
GTT |
V |
292 |
193 |
48 |
7 |
3 |
2 |
14 |
3 |
4 |
1 |
6 |
2 |
| 13 |
GTC |
V |
81 |
43 |
21 |
1 |
0 |
3 |
3 |
0 |
1 |
0 |
0 |
0 |
| 14 |
GTA |
V |
137 |
78 |
15 |
3 |
6 |
1 |
7 |
0 |
2 |
0 |
4 |
2 |
| 15 |
GTG |
V |
88 |
57 |
13 |
2 |
3 |
2 |
1 |
0 |
1 |
0 |
2 |
0 |
| 16 |
TCT |
S |
154 |
98 |
37 |
4 |
2 |
8 |
3 |
2 |
3 |
0 |
2 |
1 |
| 17 |
TCC |
S |
27 |
21 |
12 |
0 |
3 |
3 |
4 |
1 |
0 |
0 |
1 |
0 |
| 18 |
TCA |
S |
128 |
82 |
26 |
1 |
3 |
9 |
8 |
1 |
3 |
2 |
3 |
0 |
| 19 |
TCG |
S |
5 |
5 |
2 |
1 |
1 |
2 |
0 |
0 |
0 |
0 |
1 |
0 |
| 20 |
AGT |
S |
119 |
76 |
17 |
1 |
4 |
9 |
5 |
0 |
0 |
0 |
2 |
1 |
| 21 |
AGC |
S |
23 |
12 |
5 |
1 |
2 |
6 |
2 |
0 |
1 |
0 |
0 |
0 |
| 22 |
CCT |
P |
136 |
86 |
29 |
2 |
1 |
8 |
7 |
0 |
4 |
0 |
4 |
0 |
| 23 |
CCC |
P |
17 |
11 |
4 |
0 |
0 |
7 |
0 |
0 |
0 |
0 |
1 |
0 |
| 24 |
CCA |
P |
111 |
63 |
25 |
0 |
3 |
11 |
3 |
1 |
2 |
0 |
1 |
0 |
| 25 |
CCG |
P |
10 |
4 |
0 |
0 |
1 |
2 |
2 |
0 |
0 |
0 |
1 |
1 |
| 26 |
ACT |
T |
232 |
158 |
44 |
1 |
5 |
16 |
13 |
3 |
3 |
1 |
2 |
0 |
| 27 |
ACC |
T |
47 |
33 |
10 |
0 |
3 |
6 |
2 |
0 |
0 |
0 |
0 |
0 |
| 28 |
ACA |
T |
224 |
142 |
40 |
2 |
3 |
8 |
6 |
0 |
7 |
0 |
3 |
1 |
| 29 |
ACG |
T |
24 |
12 |
3 |
1 |
2 |
2 |
3 |
0 |
0 |
0 |
0 |
1 |
| 30 |
GCT |
A |
269 |
166 |
42 |
1 |
12 |
19 |
7 |
0 |
4 |
0 |
3 |
1 |
| 31 |
GCC |
A |
71 |
47 |
8 |
1 |
2 |
7 |
3 |
0 |
1 |
2 |
0 |
0 |
| 32 |
GCA |
A |
131 |
83 |
27 |
0 |
4 |
8 |
3 |
1 |
3 |
0 |
2 |
1 |
| 33 |
GCG |
A |
16 |
13 |
2 |
2 |
1 |
3 |
0 |
0 |
1 |
0 |
0 |
0 |
| 34 |
TAT |
Y |
208 |
112 |
40 |
0 |
4 |
2 |
8 |
1 |
2 |
1 |
6 |
2 |
| 35 |
TAC |
Y |
127 |
83 |
14 |
4 |
5 |
9 |
9 |
1 |
3 |
0 |
1 |
1 |
| 36 |
CAT |
H |
102 |
53 |
13 |
0 |
4 |
3 |
4 |
1 |
1 |
1 |
2 |
0 |
| 37 |
CAC |
H |
43 |
22 |
4 |
0 |
1 |
1 |
4 |
0 |
2 |
1 |
2 |
0 |
| 38 |
CAA |
Q |
163 |
97 |
46 |
0 |
2 |
27 |
5 |
2 |
4 |
1 |
3 |
1 |
| 39 |
CAG |
Q |
76 |
54 |
16 |
0 |
2 |
8 |
4 |
1 |
1 |
0 |
3 |
0 |
| 40 |
AAT |
N |
268 |
161 |
54 |
4 |
4 |
16 |
4 |
3 |
1 |
1 |
2 |
2 |
| 41 |
AAC |
N |
116 |
72 |
34 |
1 |
7 |
6 |
4 |
1 |
1 |
0 |
0 |
3 |
| 42 |
AAA |
K |
281 |
174 |
38 |
2 |
4 |
21 |
7 |
3 |
6 |
0 |
5 |
0 |
| 43 |
AAG |
K |
153 |
102 |
23 |
0 |
3 |
10 |
4 |
1 |
1 |
0 |
0 |
0 |
| 44 |
GAT |
D |
251 |
133 |
43 |
1 |
1 |
14 |
7 |
3 |
1 |
1 |
4 |
1 |
| 45 |
GAC |
D |
138 |
78 |
19 |
0 |
5 |
10 |
6 |
1 |
1 |
1 |
3 |
0 |
| 46 |
GAA |
E |
248 |
176 |
34 |
1 |
6 |
8 |
10 |
1 |
4 |
3 |
4 |
0 |
| 47 |
GAG |
E |
92 |
63 |
14 |
1 |
1 |
4 |
1 |
4 |
4 |
0 |
2 |
0 |
| 48 |
TGT |
C |
184 |
113 |
28 |
1 |
4 |
0 |
3 |
0 |
3 |
1 |
5 |
0 |
| 49 |
TGC |
C |
42 |
25 |
12 |
2 |
0 |
0 |
4 |
0 |
3 |
1 |
2 |
1 |
| 50 |
CGT |
R |
59 |
34 |
9 |
1 |
5 |
6 |
1 |
0 |
1 |
0 |
2 |
1 |
| 51 |
CGC |
R |
25 |
12 |
1 |
0 |
2 |
5 |
1 |
0 |
0 |
0 |
0 |
0 |
| 52 |
CGA |
R |
10 |
7 |
0 |
1 |
1 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
| 53 |
CGG |
R |
7 |
3 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| 54 |
AGA |
R |
112 |
55 |
20 |
1 |
3 |
10 |
3 |
0 |
4 |
0 |
2 |
1 |
| 55 |
AGG |
R |
31 |
20 |
10 |
0 |
3 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
| 56 |
GGT |
G |
263 |
172 |
47 |
1 |
5 |
10 |
7 |
0 |
1 |
0 |
3 |
0 |
| 57 |
GGC |
G |
63 |
42 |
15 |
0 |
3 |
16 |
3 |
0 |
2 |
0 |
0 |
1 |
| 58 |
GGA |
G |
76 |
44 |
17 |
0 |
6 |
13 |
4 |
0 |
1 |
0 |
2 |
0 |
| 59 |
GGG |
G |
10 |
6 |
3 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| 60 |
ATG |
M |
168 |
105 |
14 |
1 |
4 |
7 |
4 |
3 |
1 |
2 |
1 |
2 |
| 61 |
TGG |
W |
78 |
46 |
12 |
0 |
7 |
5 |
6 |
1 |
0 |
1 |
1 |
0 |
Gene-wise Relative Synonymous Codon Usage (RSCU) of SARS-CoV-2 (2019-nCoV)
| No |
codon |
AA |
ORF1ab |
ORF1a |
S |
E |
M |
N |
ORF3a |
ORF6 |
ORF7a |
ORF7b |
ORF8 |
ORF10 |
| 1 |
TTT |
F |
1.46 |
1.5 |
1.532 |
0.8 |
0.909 |
0.462 |
1.143 |
2 |
1.4 |
1 |
1 |
1.5 |
| 2 |
TTC |
F |
0.54 |
0.5 |
0.468 |
1.2 |
1.091 |
1.538 |
0.857 |
0 |
0.6 |
1 |
1 |
0.5 |
| 3 |
TTA |
L |
1.81 |
1.614 |
1.556 |
0.429 |
0.686 |
0.444 |
0.6 |
2.25 |
0.8 |
1.636 |
3.6 |
0 |
| 4 |
TTG |
L |
1.04 |
1.2 |
1.111 |
0.857 |
0.686 |
2 |
1.8 |
0 |
0.4 |
0.545 |
1.2 |
1.5 |
| 5 |
CTT |
L |
1.66 |
1.779 |
2 |
3 |
2.057 |
1.778 |
2 |
0.75 |
2.4 |
2.182 |
1.2 |
0 |
| 6 |
CTC |
L |
0.55 |
0.538 |
0.667 |
0 |
1.029 |
0.444 |
1 |
1.5 |
0.8 |
0 |
0 |
3 |
| 7 |
CTA |
L |
0.69 |
0.621 |
0.5 |
0.857 |
0.857 |
0.667 |
0.2 |
1.5 |
0.4 |
0.545 |
0 |
1.5 |
| 8 |
CTG |
L |
0.25 |
0.248 |
0.167 |
0.857 |
0.686 |
0.667 |
0.4 |
0 |
1.2 |
1.091 |
0 |
0 |
| 9 |
ATT |
I |
1.48 |
1.521 |
1.737 |
1 |
1.65 |
1.929 |
1.286 |
1.5 |
1.5 |
2.4 |
1.5 |
0 |
| 10 |
ATC |
I |
0.5 |
0.53 |
0.553 |
1 |
0.9 |
0.857 |
0.714 |
0.3 |
0.375 |
0.6 |
1.5 |
0 |
| 11 |
ATA |
I |
1.02 |
0.949 |
0.711 |
1 |
0.45 |
0.214 |
1 |
1.2 |
1.125 |
0 |
0 |
3 |
| 12 |
GTT |
V |
1.95 |
2.081 |
1.979 |
2.154 |
1 |
1 |
2.24 |
4 |
2 |
4 |
2 |
2 |
| 13 |
GTC |
V |
0.54 |
0.464 |
0.866 |
0.308 |
0 |
1.5 |
0.48 |
0 |
0.5 |
0 |
0 |
0 |
| 14 |
GTA |
V |
0.92 |
0.841 |
0.619 |
0.923 |
2 |
0.5 |
1.12 |
0 |
1 |
0 |
1.333 |
2 |
| 15 |
GTG |
V |
0.59 |
0.615 |
0.536 |
0.615 |
1 |
1 |
0.16 |
0 |
0.5 |
0 |
0.667 |
0 |
| 16 |
TCT |
S |
2.03 |
2 |
2.242 |
3 |
0.8 |
1.297 |
0.818 |
3 |
2.571 |
0 |
1.333 |
3 |
| 17 |
TCC |
S |
0.36 |
0.429 |
0.727 |
0 |
1.2 |
0.486 |
1.091 |
1.5 |
0 |
0 |
0.667 |
0 |
| 18 |
TCA |
S |
1.68 |
1.673 |
1.576 |
0.75 |
1.2 |
1.459 |
2.182 |
1.5 |
2.571 |
6 |
2 |
0 |
| 19 |
TCG |
S |
0.07 |
0.102 |
0.121 |
0.75 |
0.4 |
0.324 |
0 |
0 |
0 |
0 |
0.667 |
0 |
| 20 |
AGT |
S |
1.57 |
1.551 |
1.03 |
0.75 |
1.6 |
1.459 |
1.364 |
0 |
0 |
0 |
1.333 |
3 |
| 21 |
AGC |
S |
0.3 |
0.245 |
0.303 |
0.75 |
0.8 |
0.973 |
0.545 |
0 |
0.857 |
0 |
0 |
0 |
| 22 |
CCT |
P |
1.99 |
2.098 |
2 |
4 |
0.8 |
1.143 |
2.333 |
0 |
2.667 |
0 |
2.286 |
0 |
| 23 |
CCC |
P |
0.25 |
0.268 |
0.276 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0.571 |
0 |
| 24 |
CCA |
P |
1.62 |
1.537 |
1.724 |
0 |
2.4 |
1.571 |
1 |
4 |
1.333 |
0 |
0.571 |
0 |
| 25 |
CCG |
P |
0.15 |
0.098 |
0 |
0 |
0.8 |
0.286 |
0.667 |
0 |
0 |
0 |
0.571 |
4 |
| 26 |
ACT |
T |
1.76 |
1.832 |
1.814 |
1 |
1.538 |
2 |
2.167 |
4 |
1.2 |
4 |
1.6 |
0 |
| 27 |
ACC |
T |
0.36 |
0.383 |
0.412 |
0 |
0.923 |
0.75 |
0.333 |
0 |
0 |
0 |
0 |
0 |
| 28 |
ACA |
T |
1.7 |
1.646 |
1.649 |
2 |
0.923 |
1 |
1 |
0 |
2.8 |
0 |
2.4 |
2 |
| 29 |
ACG |
T |
0.18 |
0.139 |
0.124 |
1 |
0.615 |
0.25 |
0.5 |
0 |
0 |
0 |
0 |
2 |
| 30 |
GCT |
A |
2.21 |
2.149 |
2.127 |
1 |
2.526 |
2.054 |
2.154 |
0 |
1.778 |
0 |
2.4 |
2 |
| 31 |
GCC |
A |
0.58 |
0.608 |
0.405 |
1 |
0.421 |
0.757 |
0.923 |
0 |
0.444 |
4 |
0 |
0 |
| 32 |
GCA |
A |
1.08 |
1.074 |
1.367 |
0 |
0.842 |
0.865 |
0.923 |
4 |
1.333 |
0 |
1.6 |
2 |
| 33 |
GCG |
A |
0.13 |
0.168 |
0.101 |
2 |
0.211 |
0.324 |
0 |
0 |
0.444 |
0 |
0 |
0 |
| 34 |
TAT |
Y |
1.24 |
1.149 |
1.481 |
0 |
0.889 |
0.364 |
0.941 |
1 |
0.8 |
2 |
1.714 |
1.333 |
| 35 |
TAC |
Y |
0.76 |
0.851 |
0.519 |
2 |
1.111 |
1.636 |
1.059 |
1 |
1.2 |
0 |
0.286 |
0.667 |
| 36 |
CAT |
H |
1.41 |
1.413 |
1.529 |
0 |
1.6 |
1.5 |
1 |
2 |
0.667 |
1 |
1 |
0 |
| 37 |
CAC |
H |
0.59 |
0.587 |
0.471 |
0 |
0.4 |
0.5 |
1 |
0 |
1.333 |
1 |
1 |
0 |
| 38 |
CAA |
Q |
1.36 |
1.285 |
1.484 |
0 |
1 |
1.543 |
1.111 |
1.333 |
1.6 |
2 |
1 |
2 |
| 39 |
CAG |
Q |
0.64 |
0.715 |
0.516 |
0 |
1 |
0.457 |
0.889 |
0.667 |
0.4 |
0 |
1 |
0 |
| 40 |
AAT |
N |
1.4 |
1.382 |
1.227 |
1.6 |
0.727 |
1.455 |
1 |
1.5 |
1 |
2 |
2 |
0.8 |
| 41 |
AAC |
N |
0.6 |
0.618 |
0.773 |
0.4 |
1.273 |
0.545 |
1 |
0.5 |
1 |
0 |
0 |
1.2 |
| 42 |
AAA |
K |
1.29 |
1.261 |
1.246 |
2 |
1.143 |
1.355 |
1.273 |
1.5 |
1.714 |
0 |
2 |
0 |
| 43 |
AAG |
K |
0.71 |
0.739 |
0.754 |
0 |
0.857 |
0.645 |
0.727 |
0.5 |
0.286 |
0 |
0 |
0 |
| 44 |
GAT |
D |
1.29 |
1.261 |
1.387 |
2 |
0.333 |
1.167 |
1.077 |
1.5 |
1 |
1 |
1.143 |
2 |
| 45 |
GAC |
D |
0.71 |
0.739 |
0.613 |
0 |
1.667 |
0.833 |
0.923 |
0.5 |
1 |
1 |
0.857 |
0 |
| 46 |
GAA |
E |
1.46 |
1.473 |
1.417 |
1 |
1.714 |
1.333 |
1.818 |
0.4 |
1 |
2 |
1.333 |
0 |
| 47 |
GAG |
E |
0.54 |
0.527 |
0.583 |
1 |
0.286 |
0.667 |
0.182 |
1.6 |
1 |
0 |
0.667 |
0 |
| 48 |
TGT |
C |
1.63 |
1.638 |
1.4 |
0.667 |
2 |
0 |
0.857 |
0 |
1 |
1 |
1.429 |
0 |
| 49 |
TGC |
C |
0.37 |
0.362 |
0.6 |
1.333 |
0 |
0 |
1.143 |
0 |
1 |
1 |
0.571 |
2 |
| 50 |
CGT |
R |
1.45 |
1.557 |
1.286 |
2 |
2.143 |
1.241 |
1 |
0 |
1.2 |
0 |
3 |
3 |
| 51 |
CGC |
R |
0.61 |
0.55 |
0.143 |
0 |
0.857 |
1.034 |
1 |
0 |
0 |
0 |
0 |
0 |
| 52 |
CGA |
R |
0.25 |
0.321 |
0 |
2 |
0.429 |
1.034 |
0 |
0 |
0 |
0 |
0 |
0 |
| 53 |
CGG |
R |
0.17 |
0.137 |
0.286 |
0 |
0 |
0.414 |
0 |
0 |
0 |
0 |
0 |
0 |
| 54 |
AGA |
R |
2.75 |
2.519 |
2.857 |
2 |
1.286 |
2.069 |
3 |
0 |
4.8 |
0 |
3 |
3 |
| 55 |
AGG |
R |
0.76 |
0.916 |
1.429 |
0 |
1.286 |
0.207 |
1 |
6 |
0 |
0 |
0 |
0 |
| 56 |
GGT |
G |
2.55 |
2.606 |
2.293 |
4 |
1.429 |
0.93 |
2 |
0 |
1 |
0 |
2.4 |
0 |
| 57 |
GGC |
G |
0.61 |
0.636 |
0.732 |
0 |
0.857 |
1.488 |
0.857 |
0 |
2 |
0 |
0 |
4 |
| 58 |
GGA |
G |
0.74 |
0.667 |
0.829 |
0 |
1.714 |
1.209 |
1.143 |
0 |
1 |
0 |
1.6 |
0 |
| 59 |
GGG |
G |
0.1 |
0.091 |
0.146 |
0 |
0 |
0.372 |
0 |
0 |
0 |
0 |
0 |
0 |
| 60 |
ATG |
M |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
| 61 |
TGG |
W |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
Codon usage, Amino acids, RSCU, Rare codons and context details of SARS-CoV-2 (2019-nCoV) genes
| | | | | |
|---|
|
| Genes | Codons | Amino acids | RSCU | Rare codons | Codon context |
| ORF1ab | 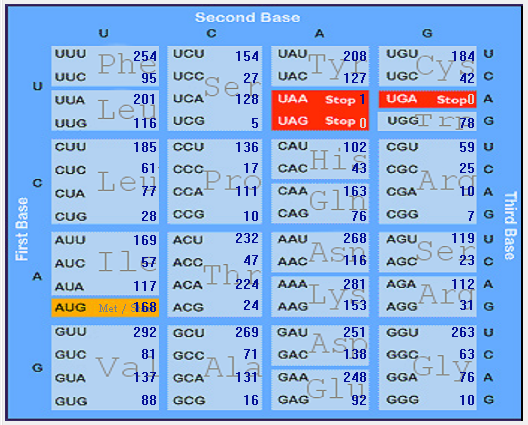 | 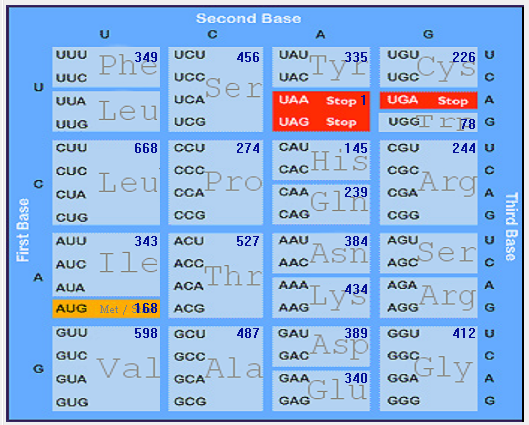 | 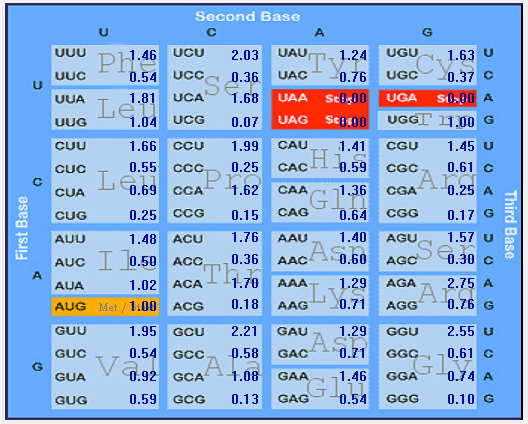 | 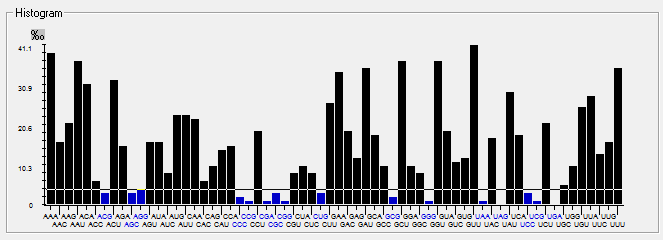 | 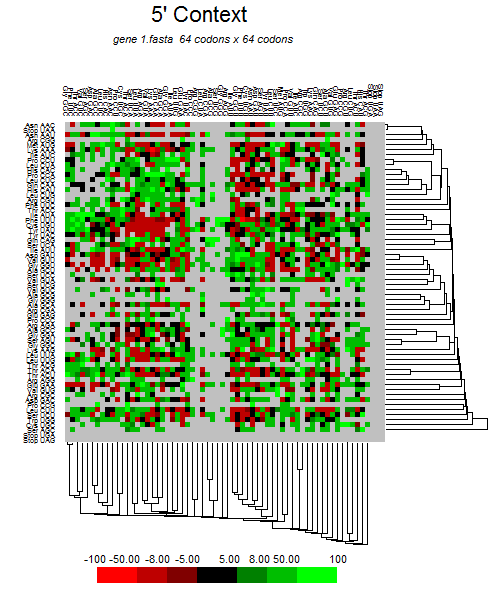 |
| ORF1a | 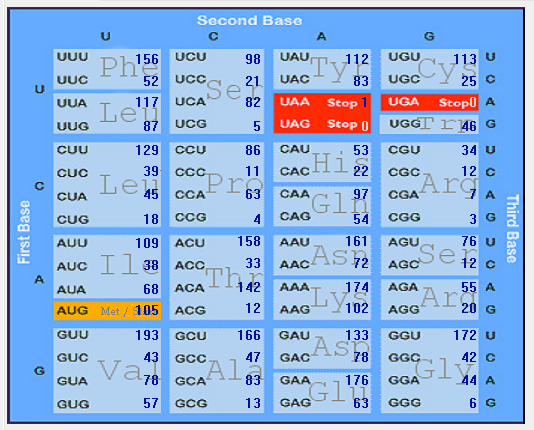 | 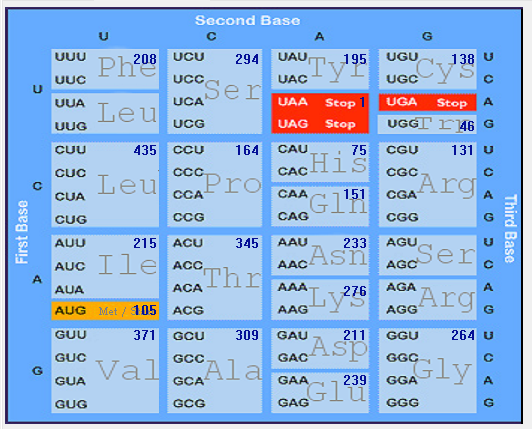 | 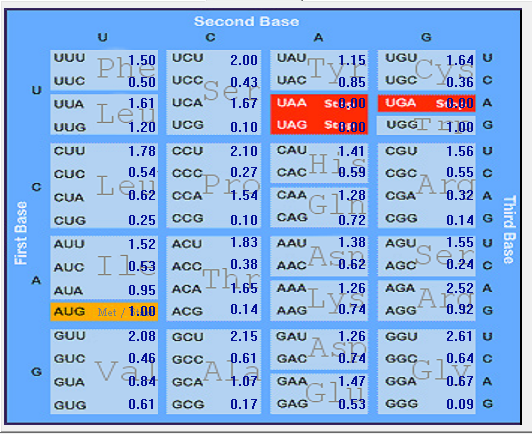 | 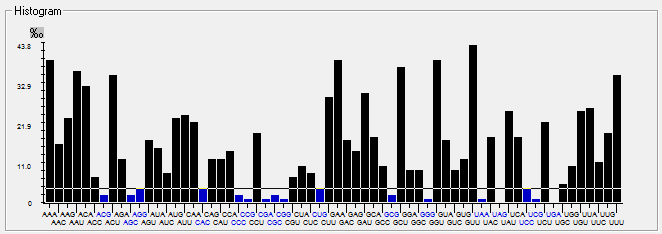 | 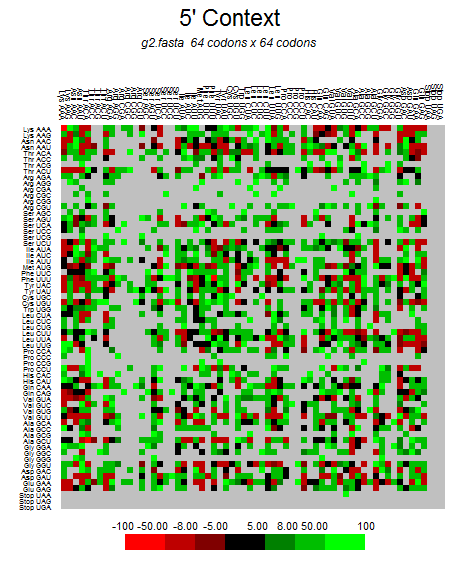 |
| S | 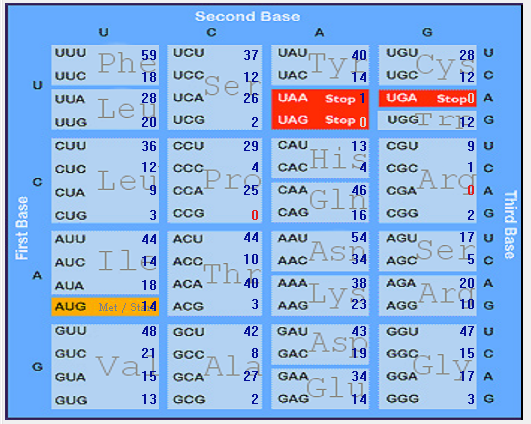 | 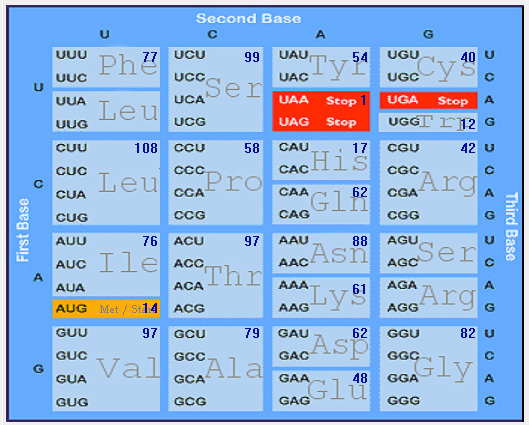 | 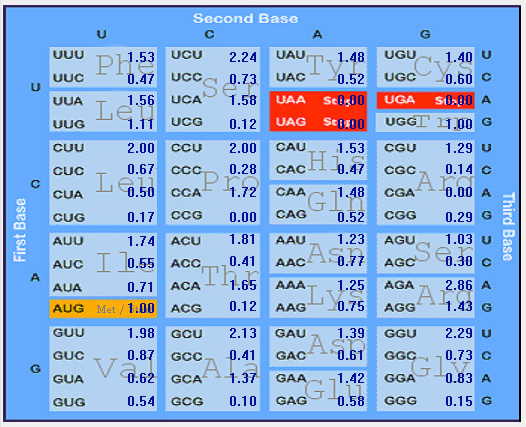 | 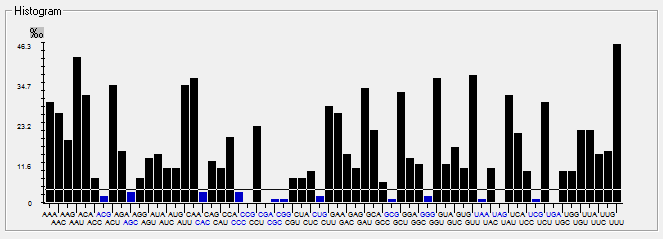 | 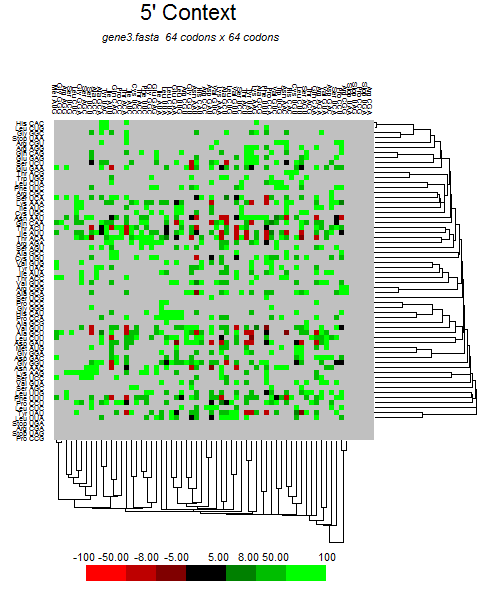 |
| E | 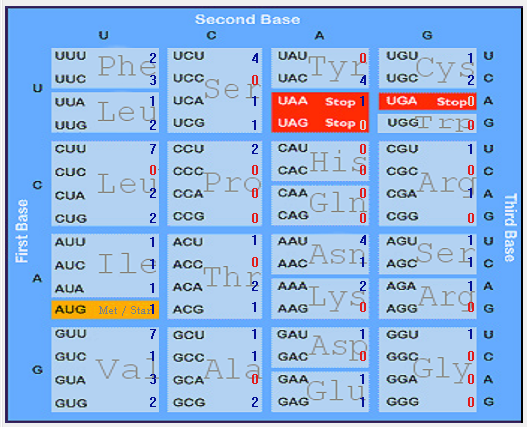 | 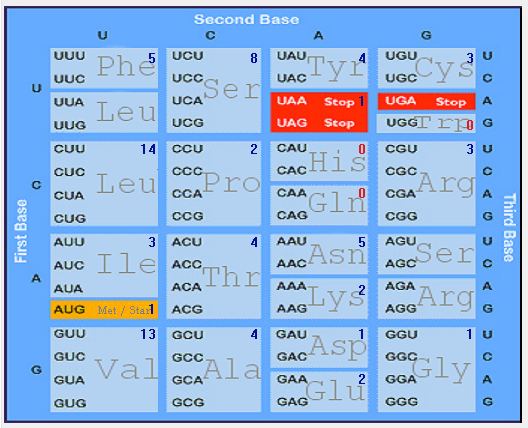 | 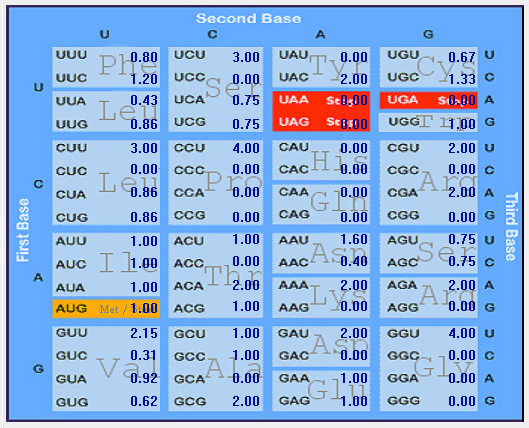 | 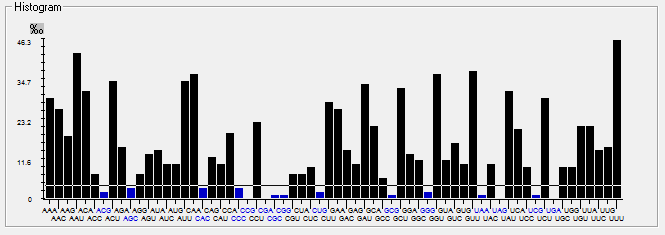 | 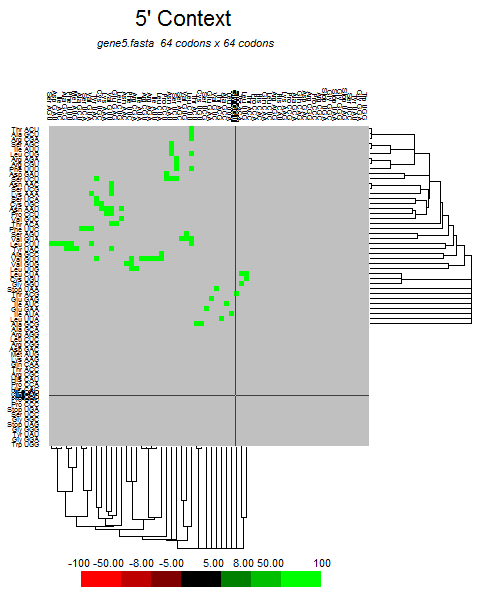 |
| M | 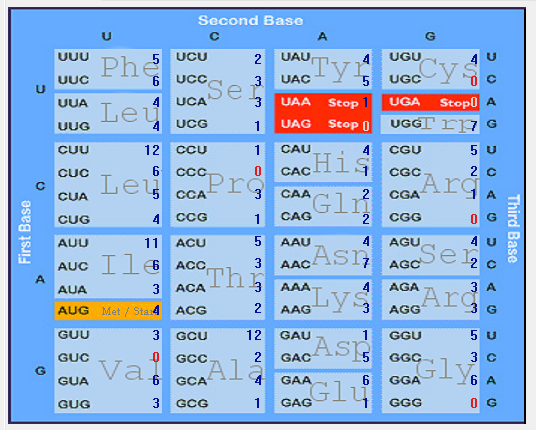 | 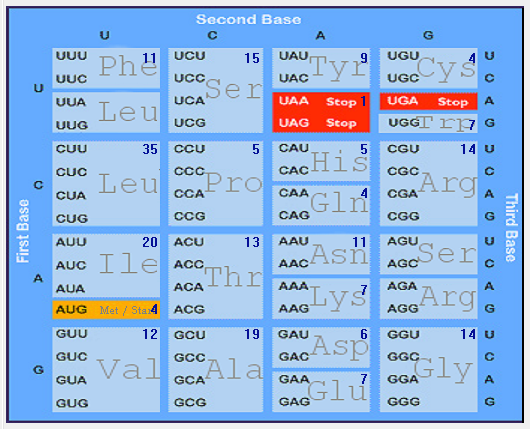 | 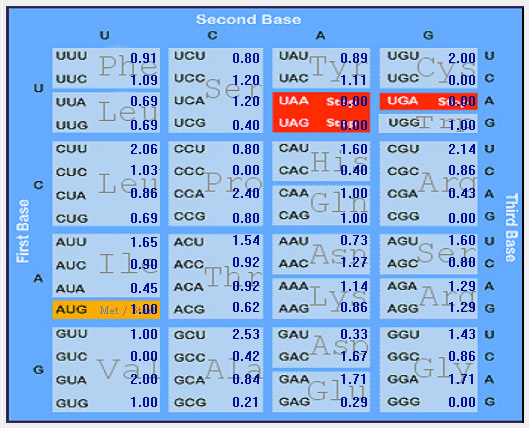 | 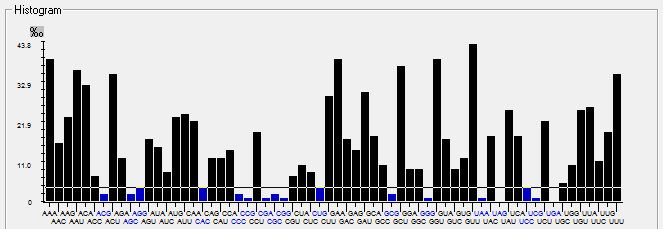 | 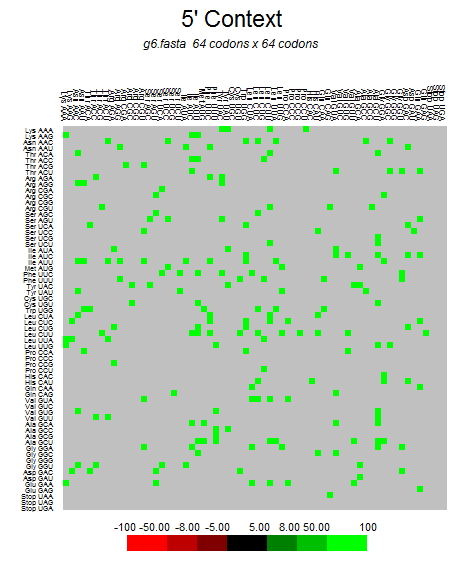 |
| N | 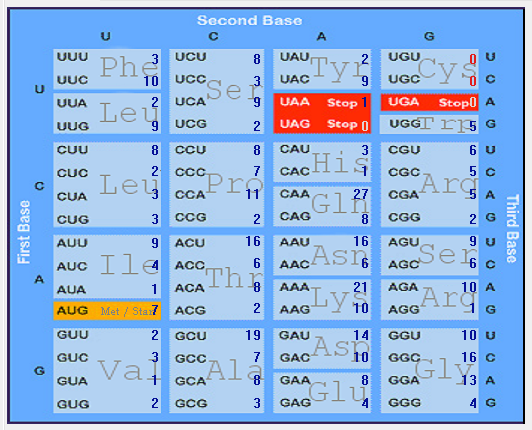 | 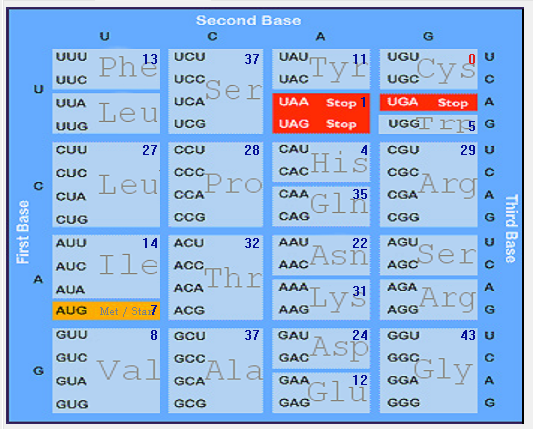 | 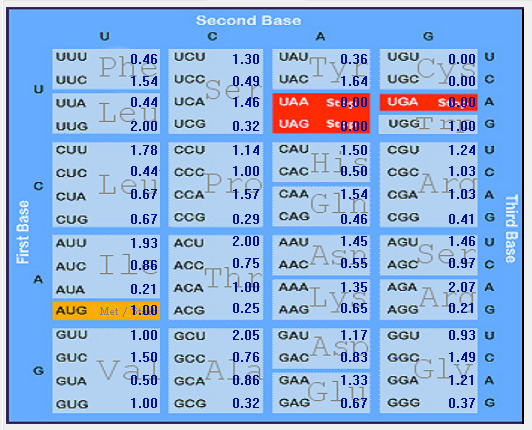 | 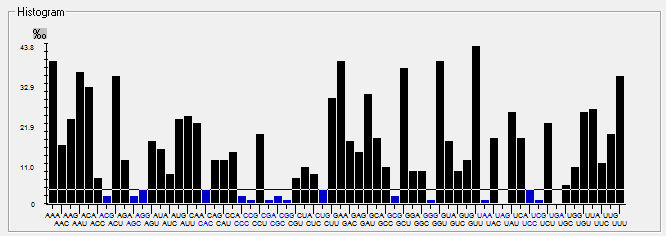 | 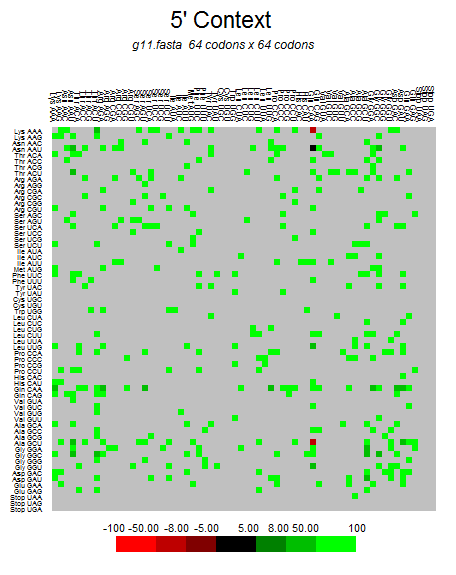 |
| ORF3a | 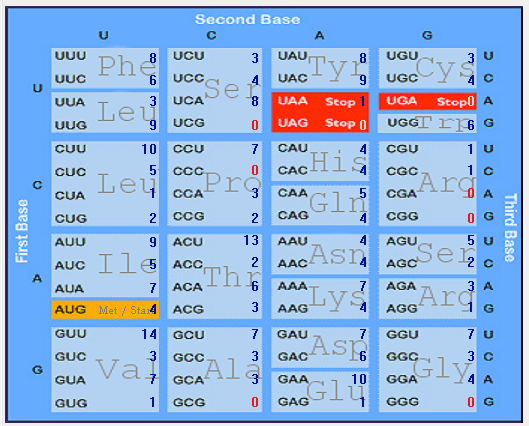 | 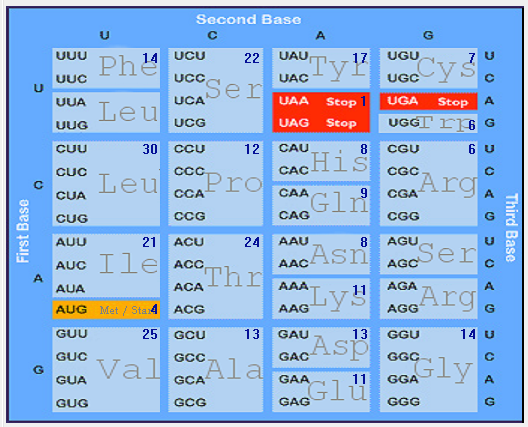 | 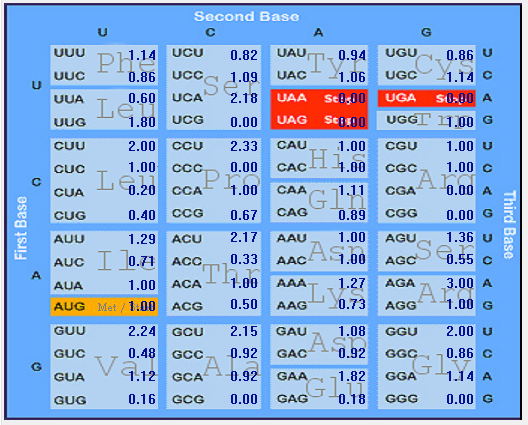 | 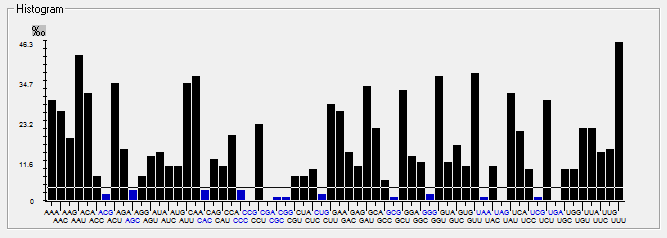 | 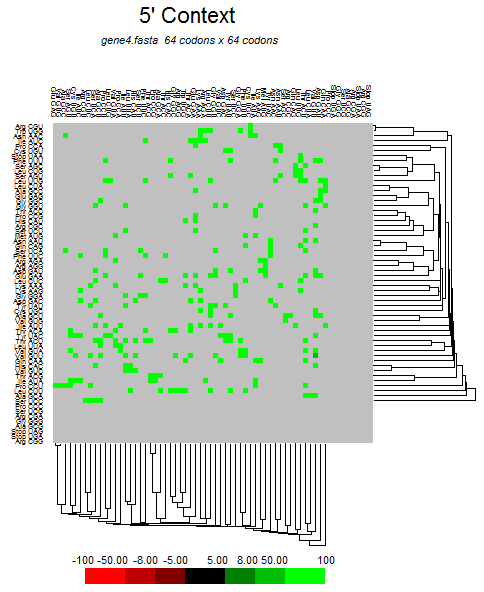 |
| ORF6 | 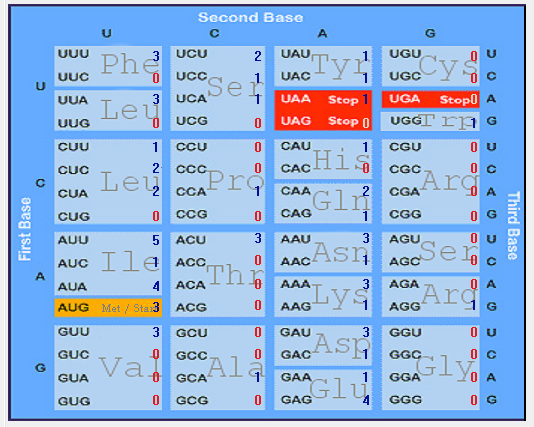 | 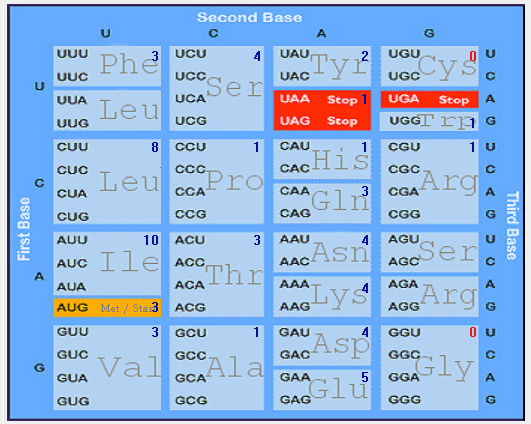 | 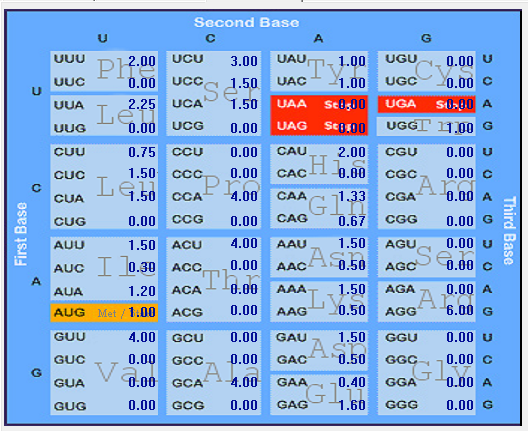 | 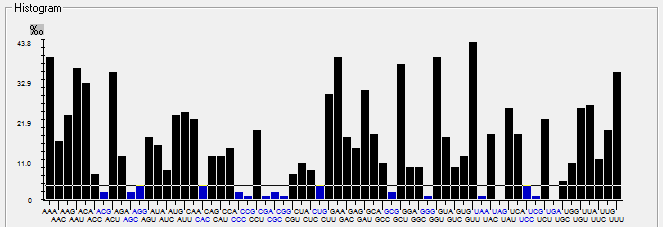 | 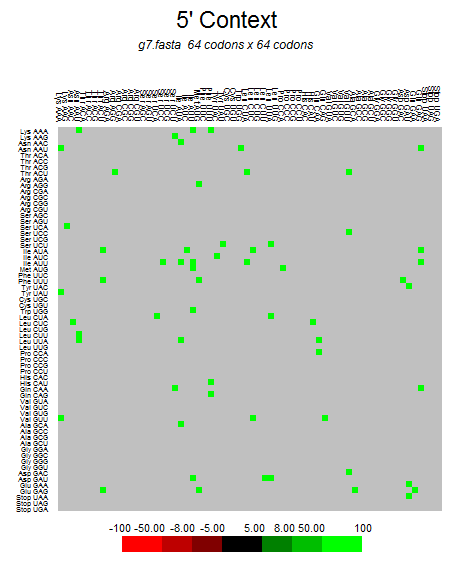 |
| ORF7a | 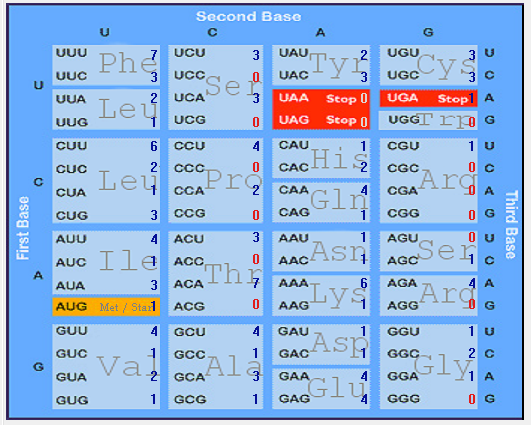 | 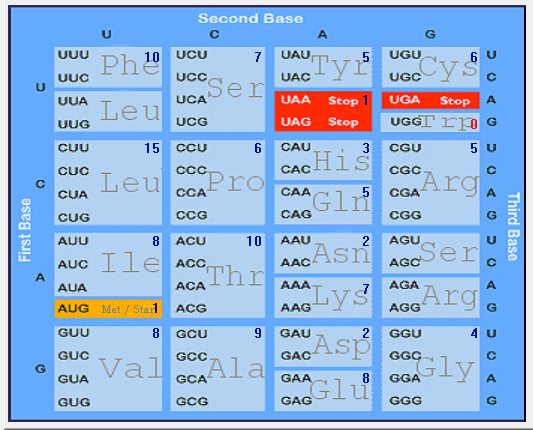 | 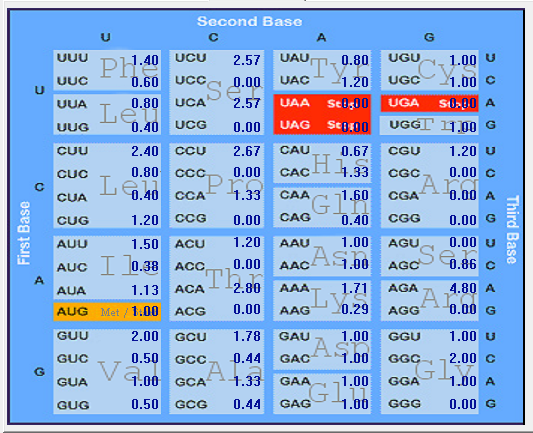 | 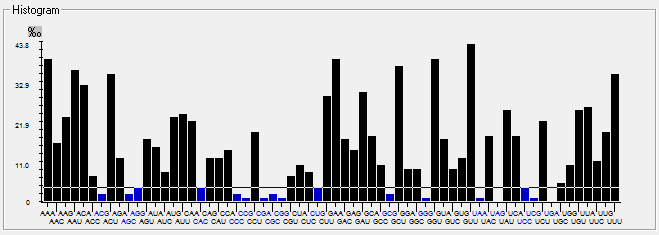 |  |
| ORF7b | 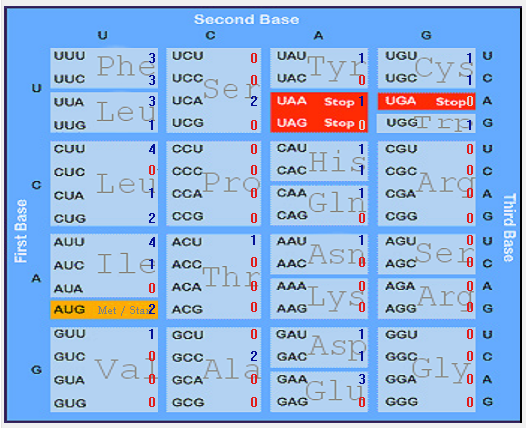 | 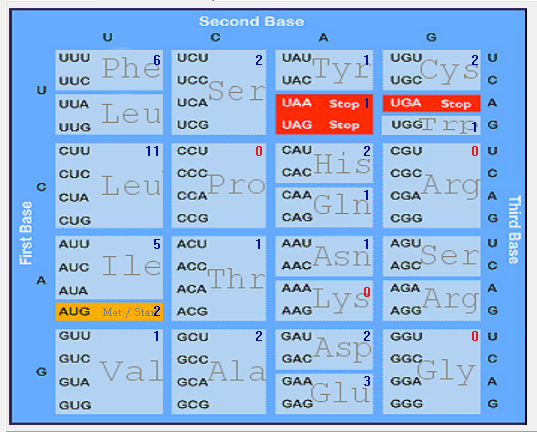 | 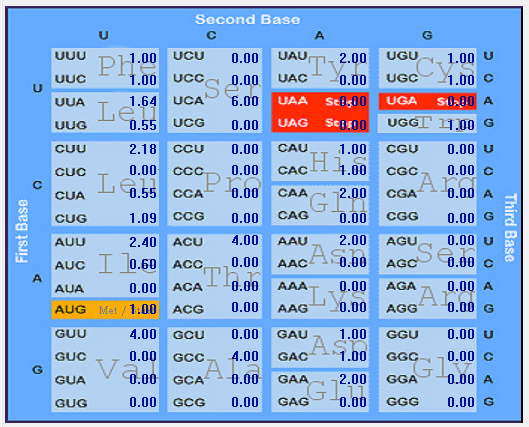 | 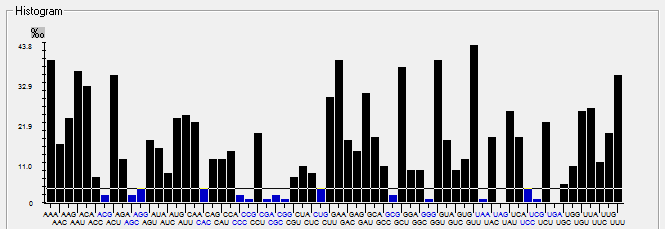 | 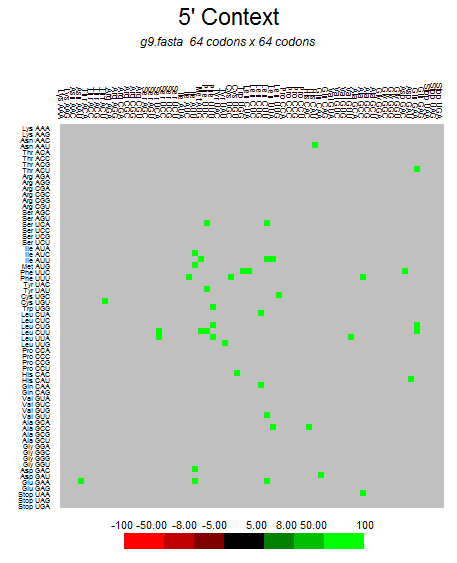 |
| ORF8 | 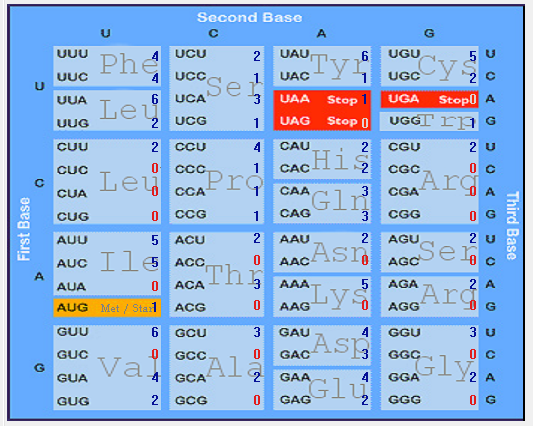 | 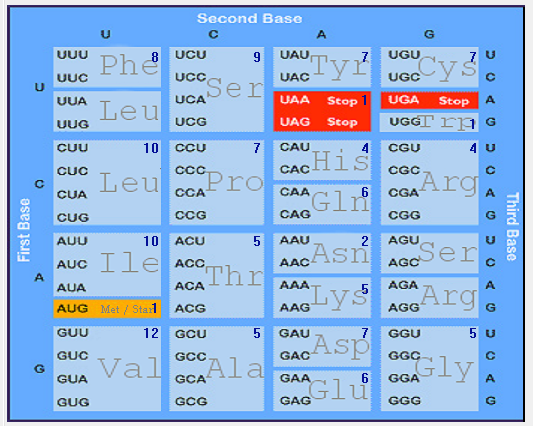 | 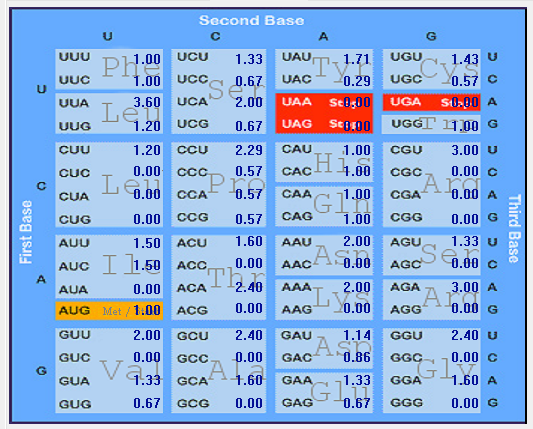 | 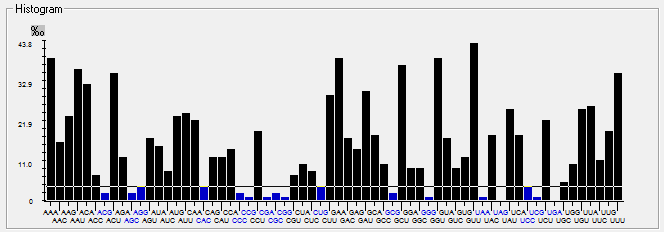 | 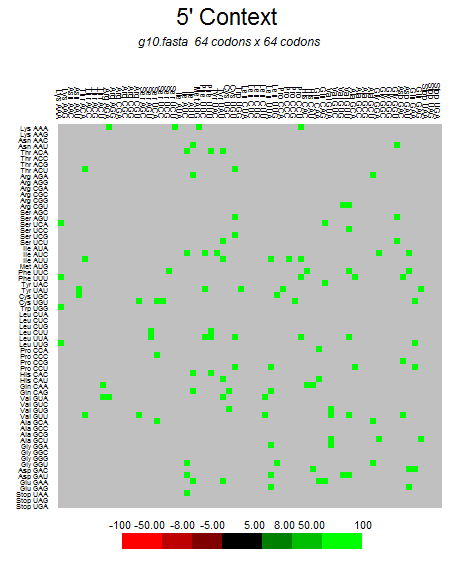 |
| ORF10 | 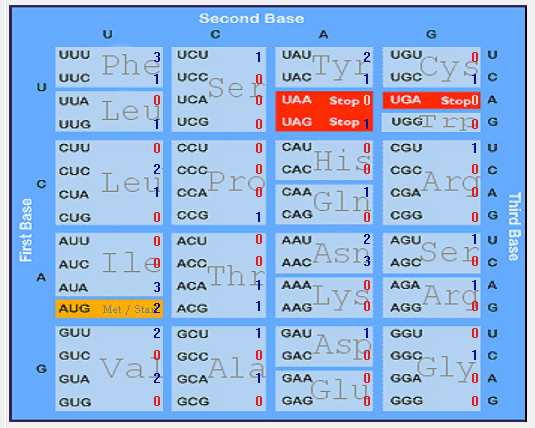 | 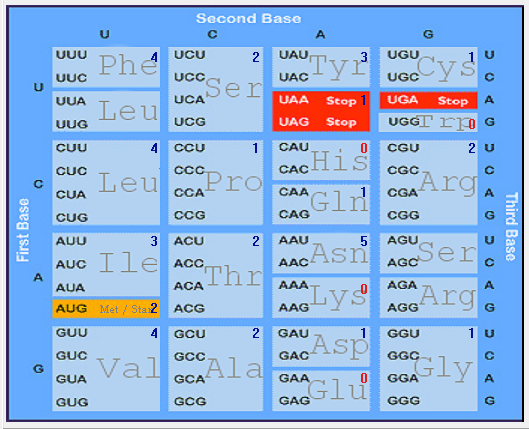 | 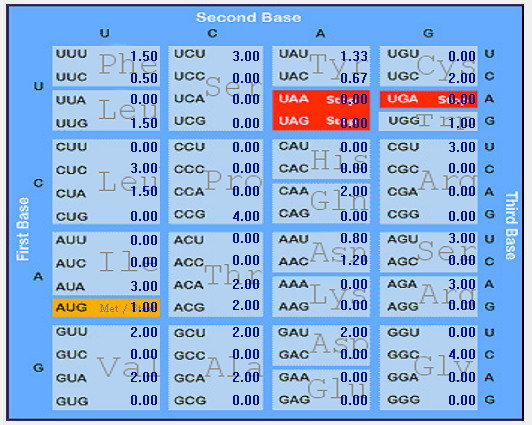 | 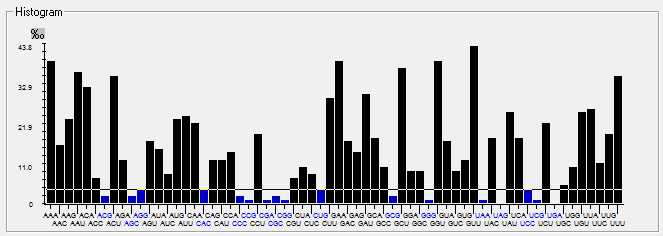 | 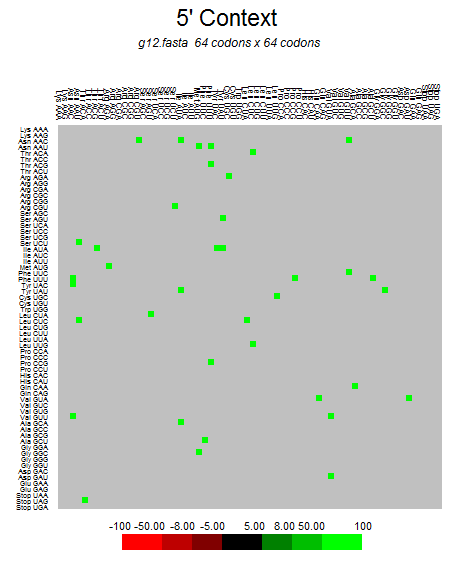 |
Genome-wise Codon usage, Amino acids, RSCU, Rare codons and context details of different coronaviruses
| | | | | |
|---|
|
| Genome | Codons | Amino acids | RSCU | Rare codons | Codon context |
| 2019-nCoV | 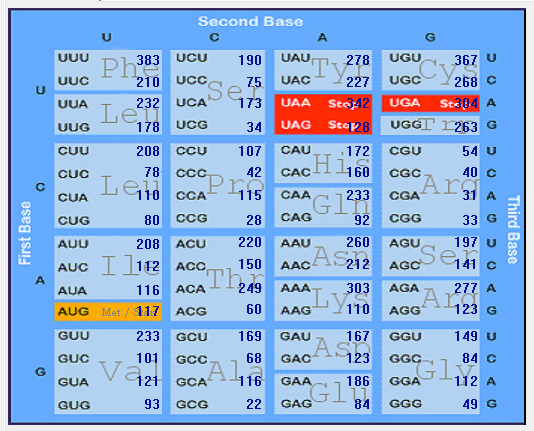 | 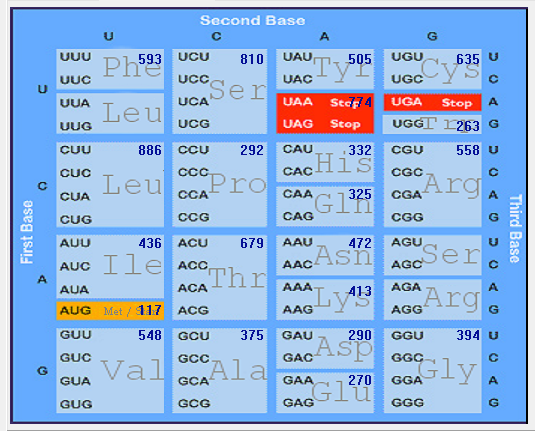 | 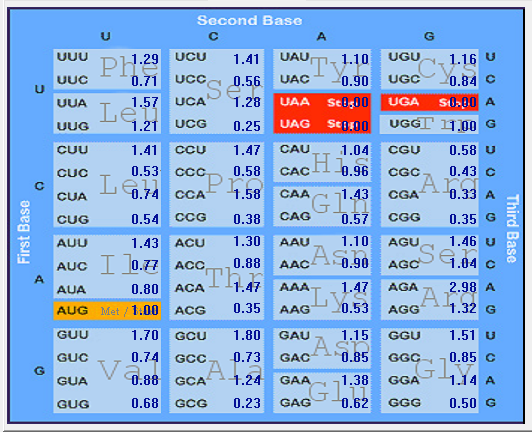 | 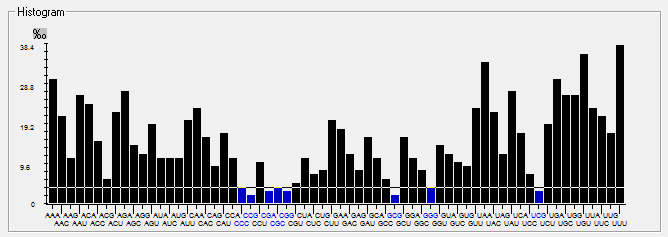 | 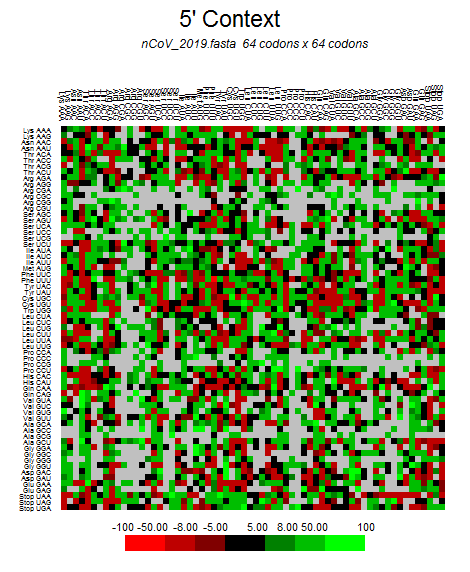 |
| SARS-CoV | 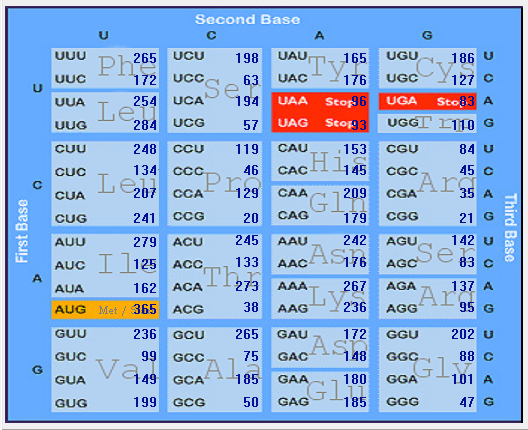 | 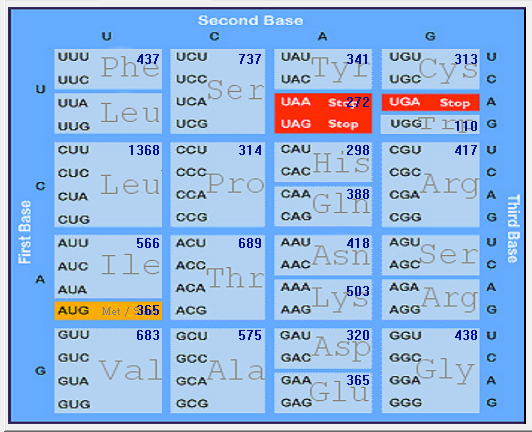 | 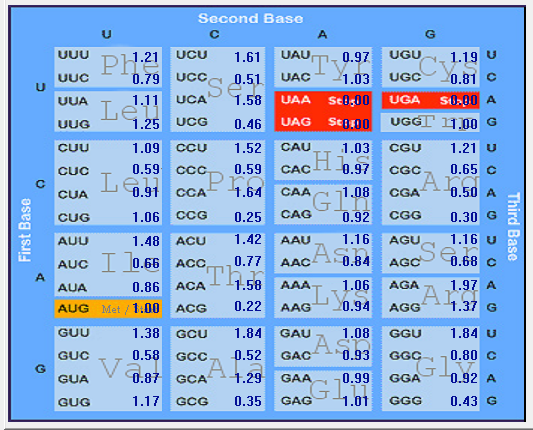 | 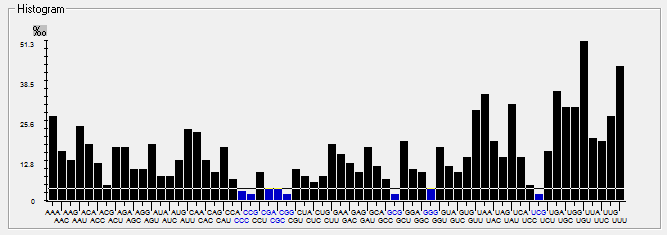 | 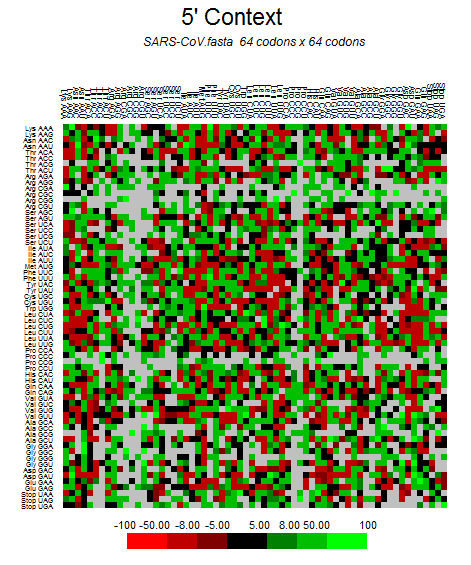 |
| MERS-CoV | 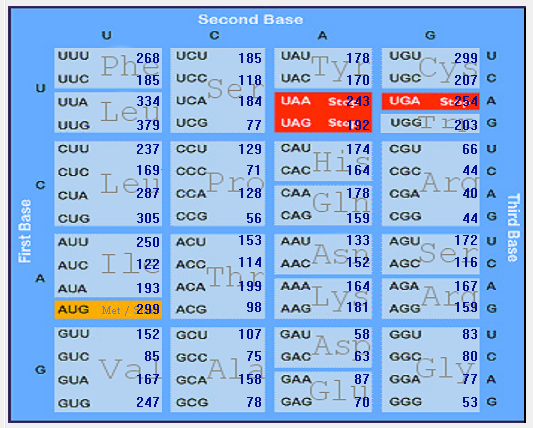 | 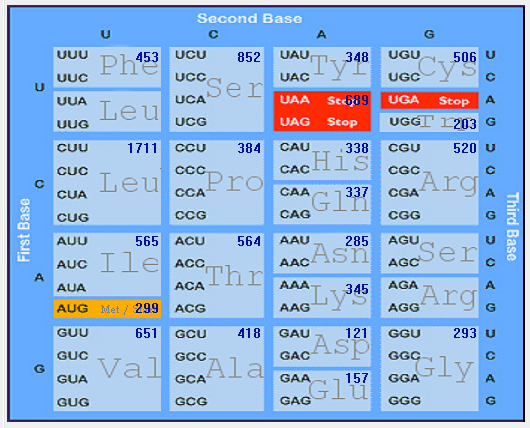 | 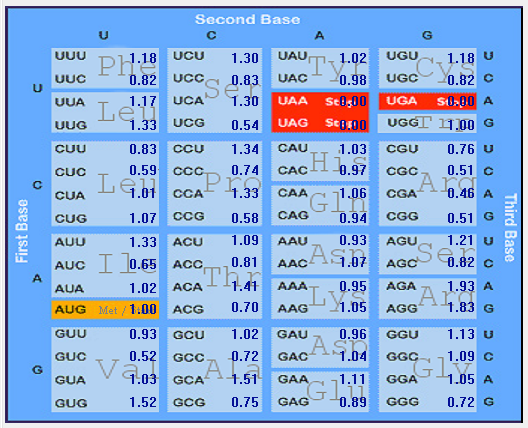 | 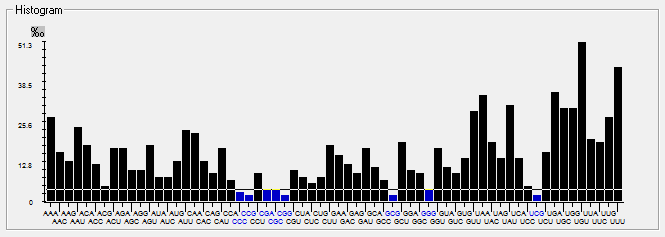 | 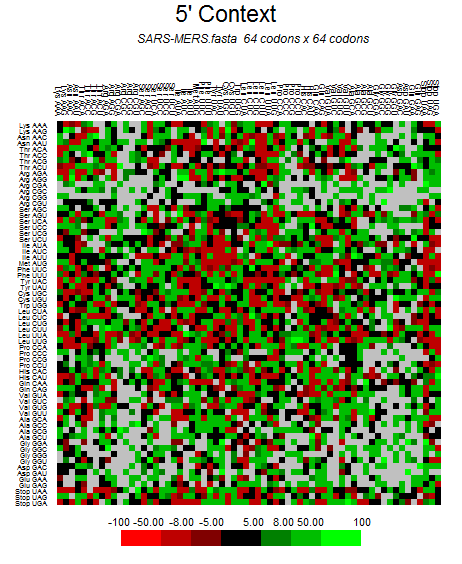 |
| HCoV-229E | 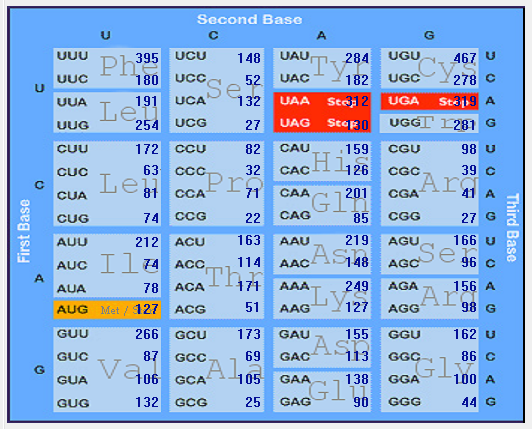 | 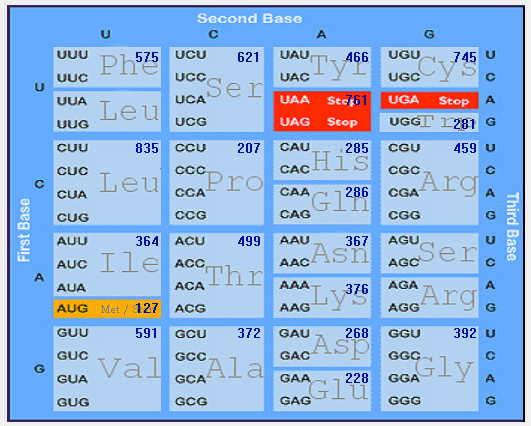 | 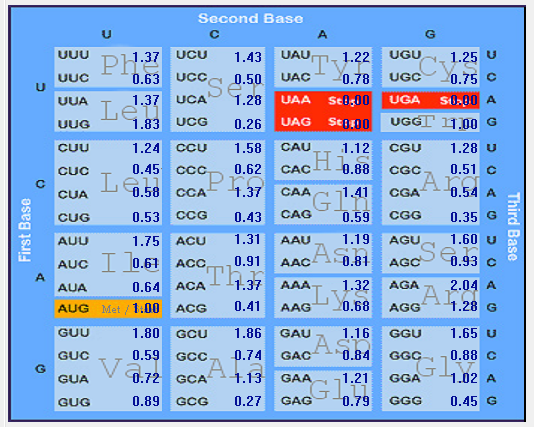 | 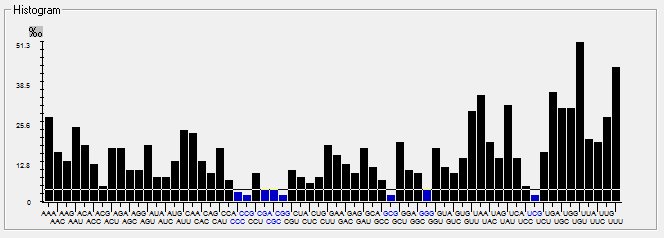 | 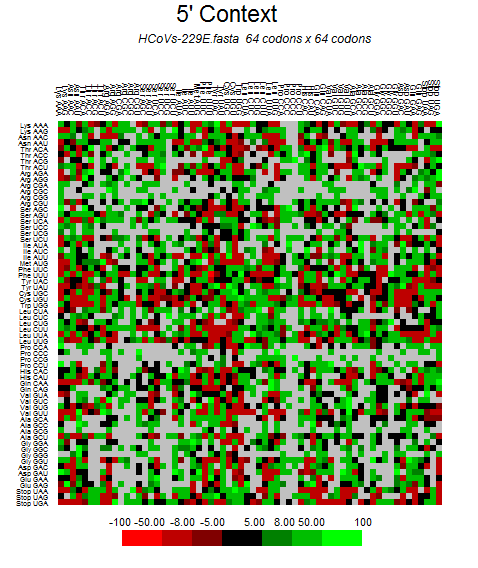 |
| HCoV-HKU1 | 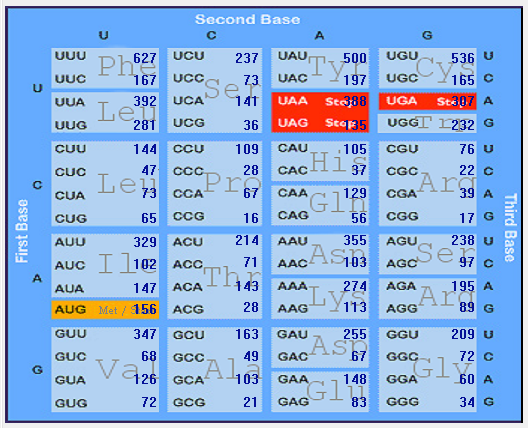 | 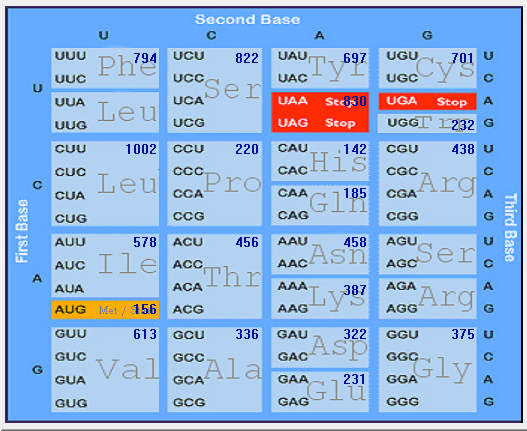 | 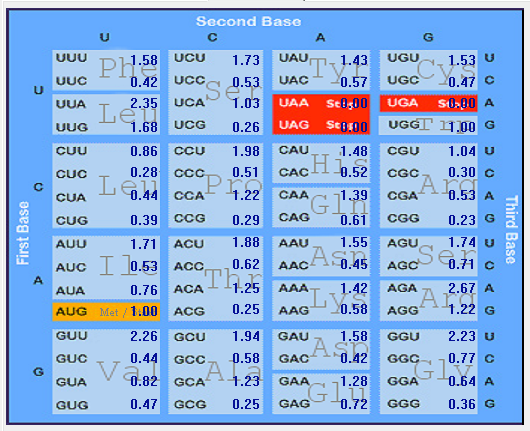 | 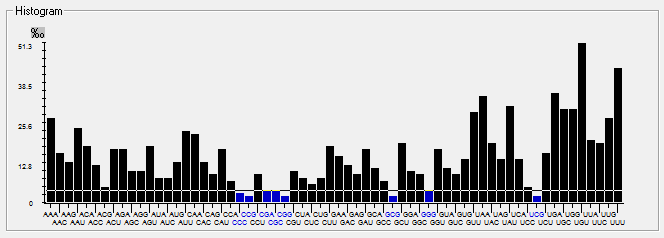 | 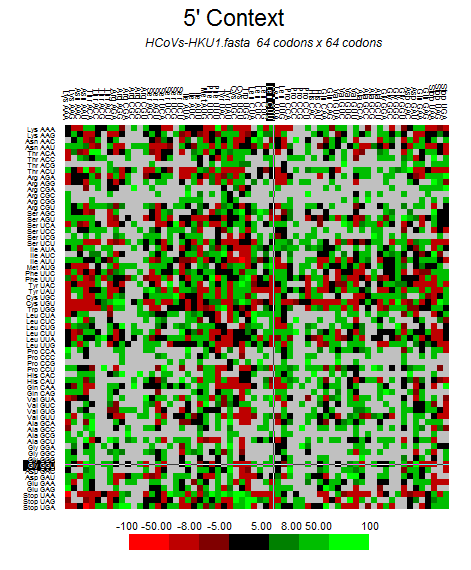 |
| HCoV-NL63 | 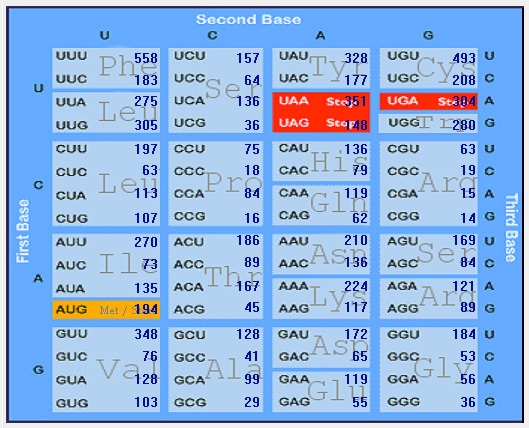 | 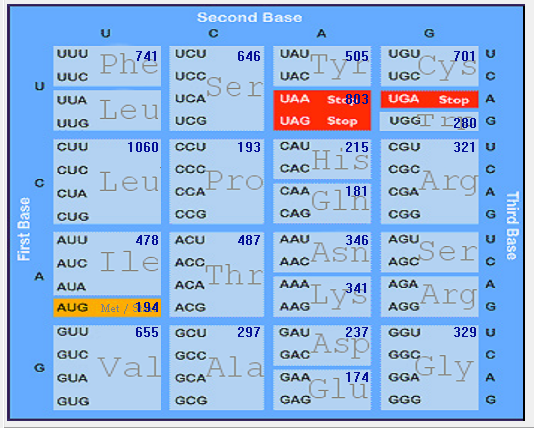 | 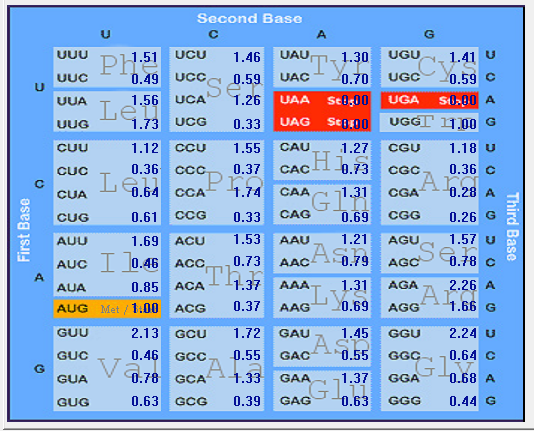 | 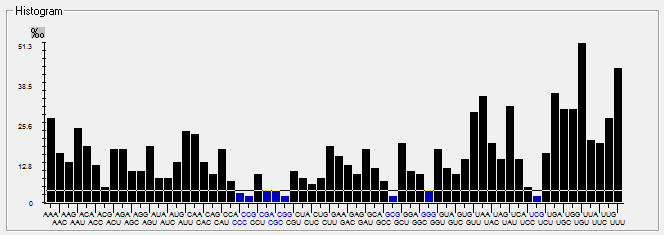 | 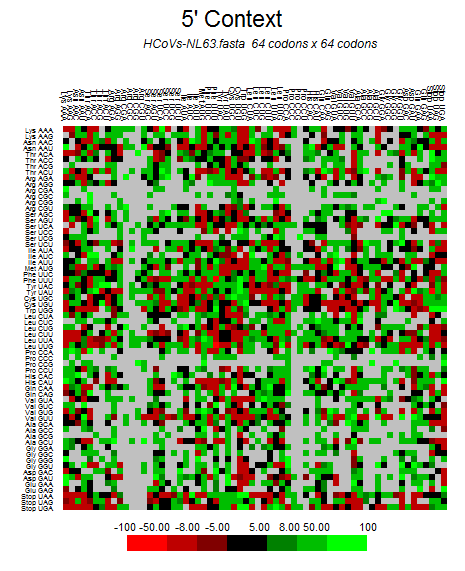 |
| HCoV-OC43 | 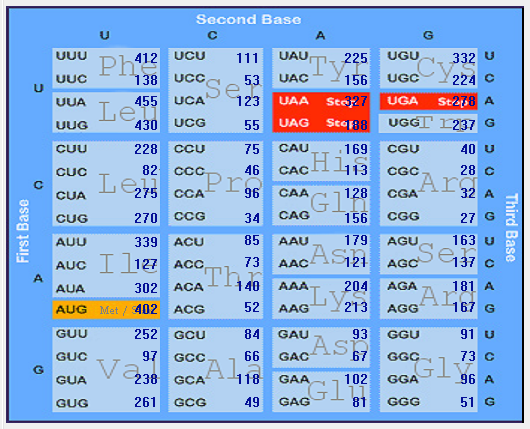 | 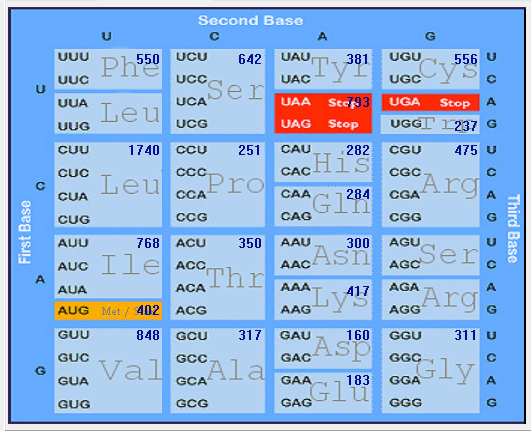 | 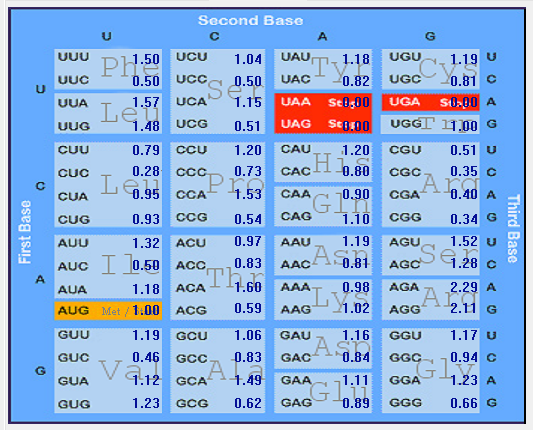 | 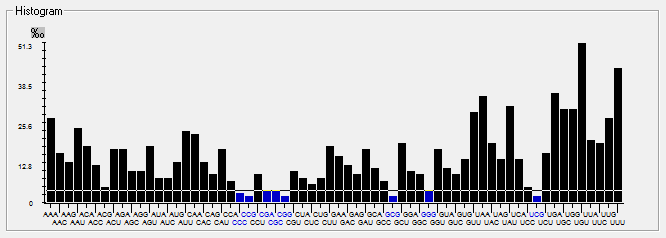 | 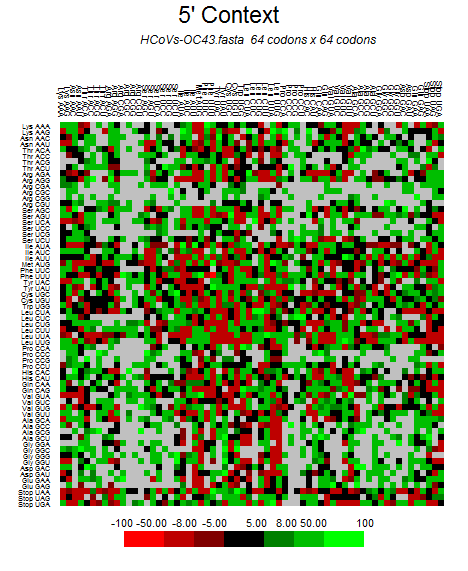 |
