How to use
ResourcevhfRNAi is a manually curated, open source resource of viral host genes experimentally validated using high throughput RNAi screen technology. Currently vhfRNAi provides information of 12249 host factors plus restriction factors belonging to 18 different human viruses. The resource is accessible at http://bioinfo.imtech.res.in/manojk/vhfrnai.
Browse
This user-friendly browse option allows you to explore the data for different viruses categorized in the database.

Search
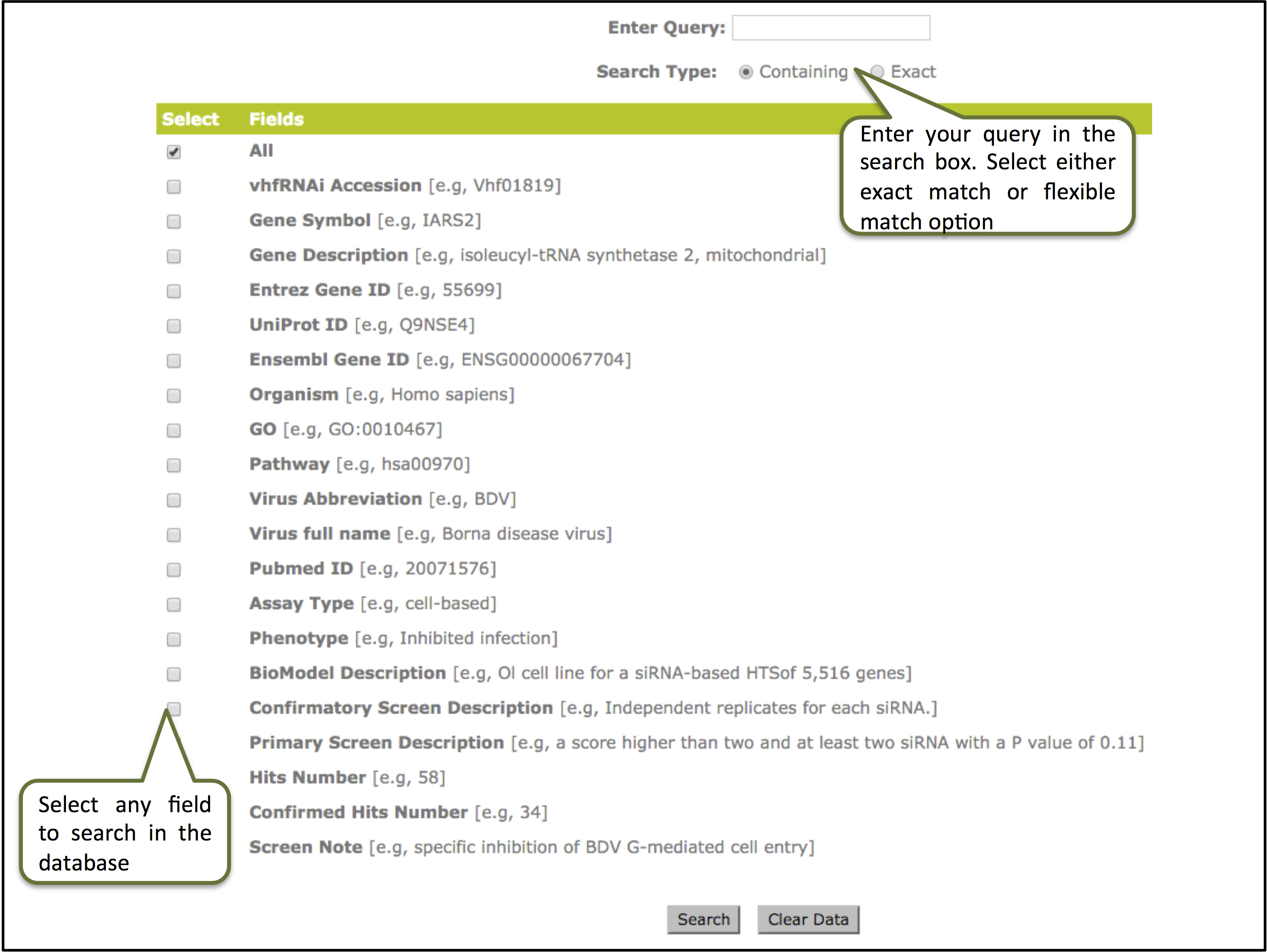
Field Search: Here user can enter the desired keyword in the given box and can execute search based on the given fields like vhfrnai_ID, gene symbol, gene description, organism, virus abbreviation or full name, biomodel description, PMID etc. User can also use the default "All" option tol search against all the fields in the database. Search type option also allow to retrieve data either match containing the query or an exact match.
Data display
The result page will display different fields of the database. The results obtained from browse/search can be sorted by clicking on the column header and filtering can be done by typing in the boxes given under the headers.
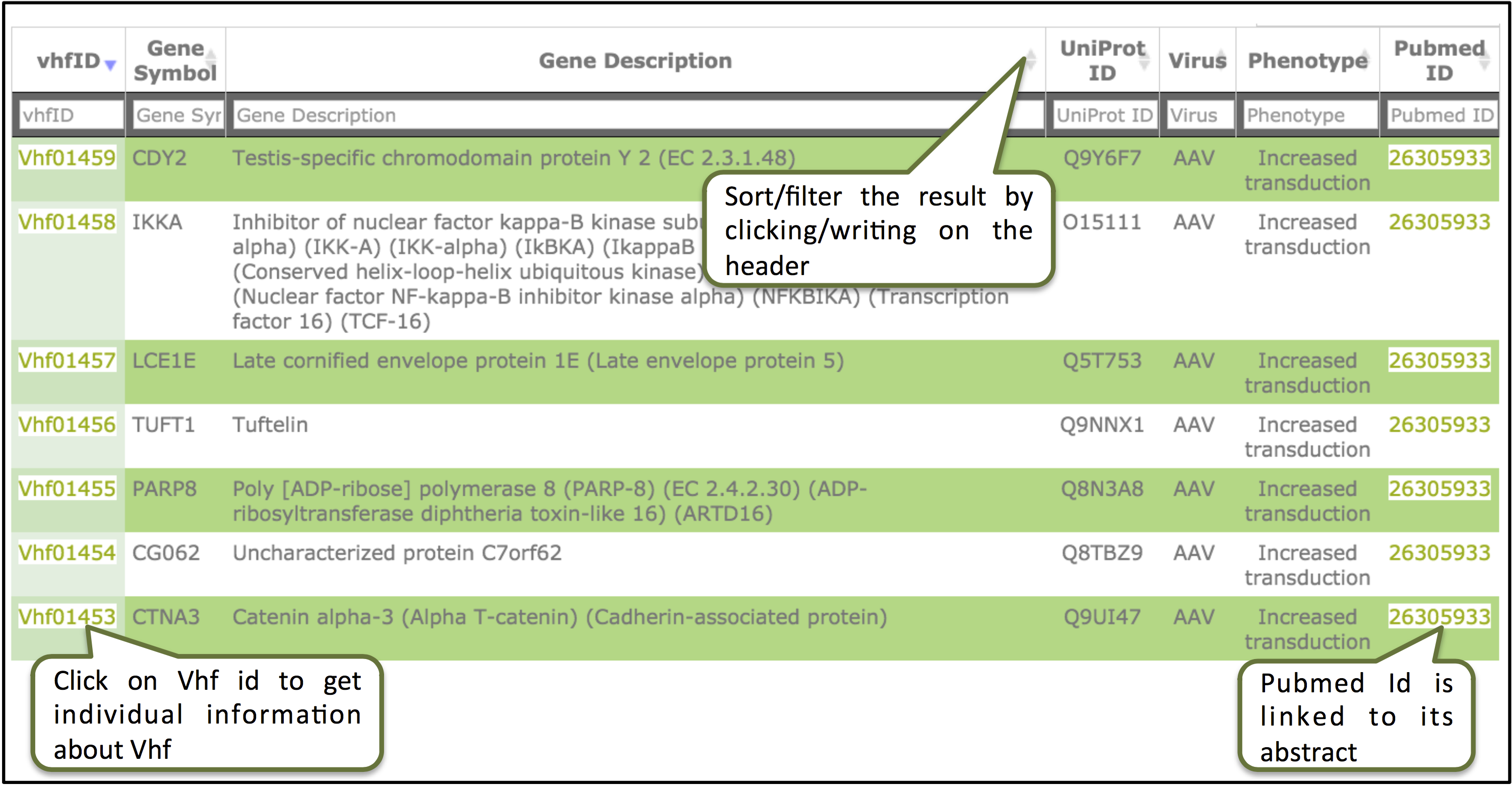
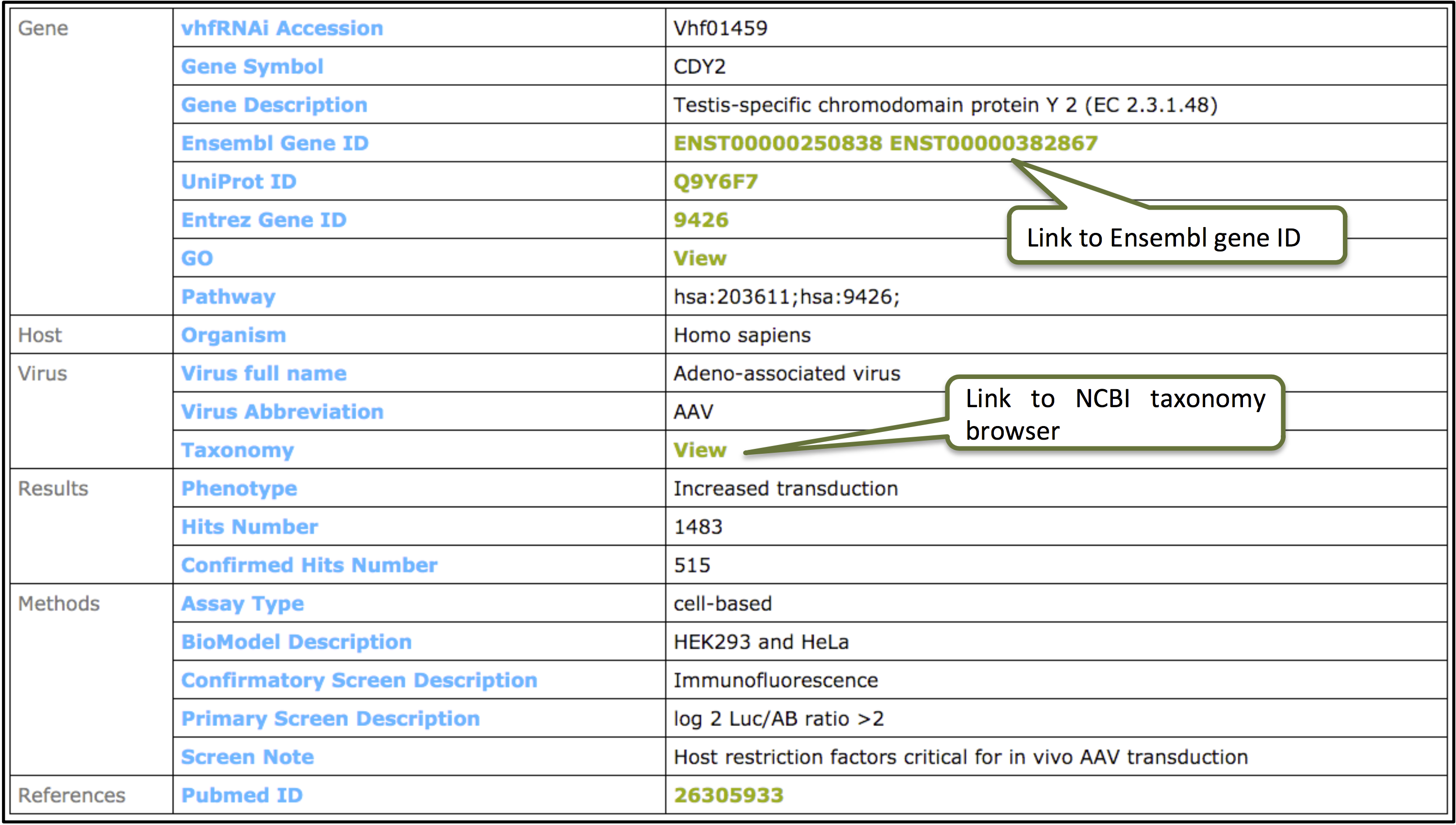
Clicking on the ID will display information for the individual record.
Gene Targeted
In gene-targeted option, user can view the list of total, neutral, up-regulated and down-regulated genes used in different studies.

Analysis tools
vhfRNAi also offers easy to use and useful tools such as network enrichment and interaction analysis to help in comprehensive understanding of genes that aid or hinder the viral propagation inside the host cell.
Interactions
The network analysis graphically depicts the connection between virus and genes (host factors + restriction factors) and their gene ontologies.
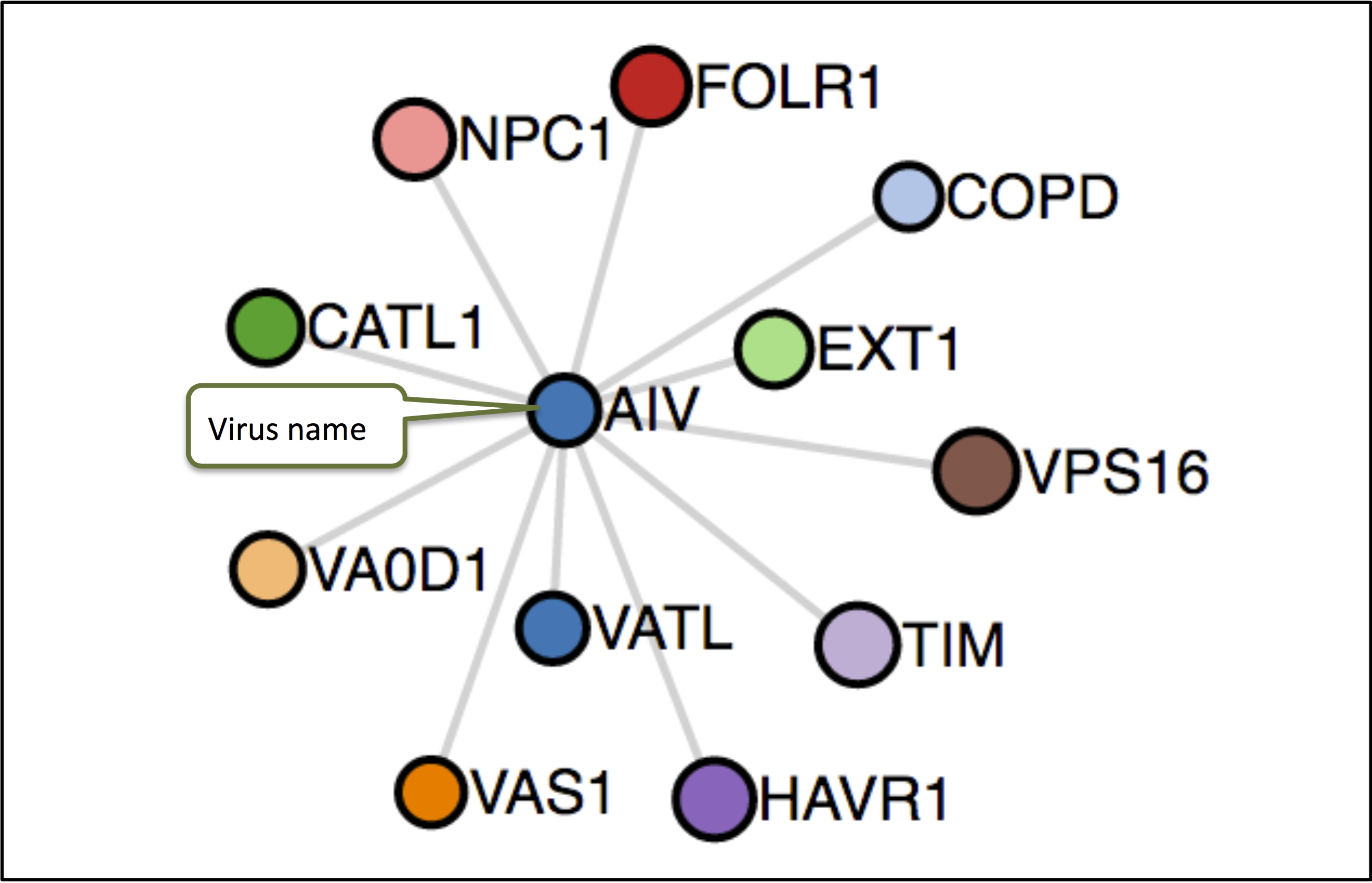
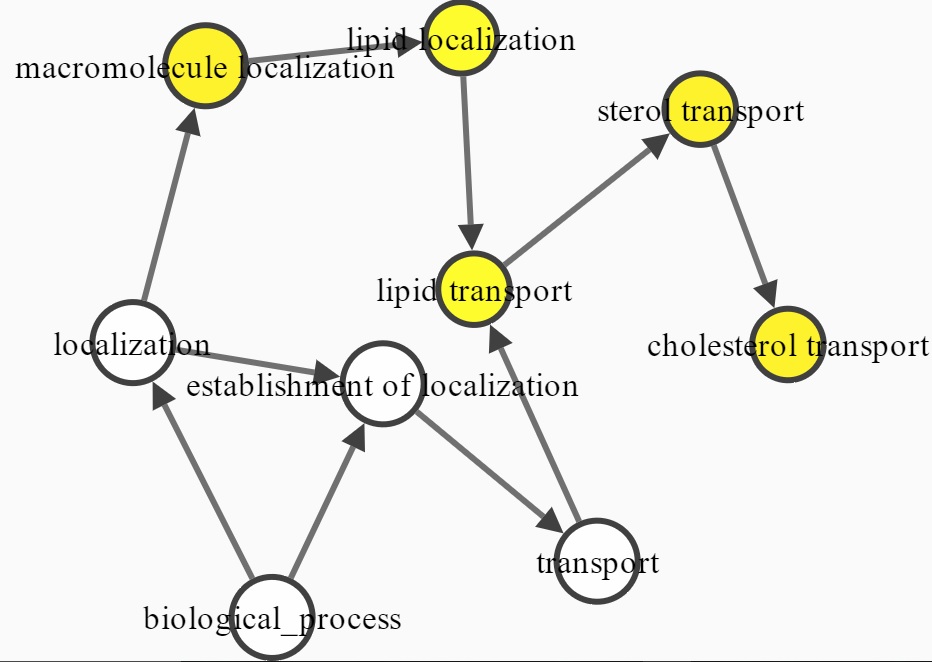
Pathway Enrichment Analysis
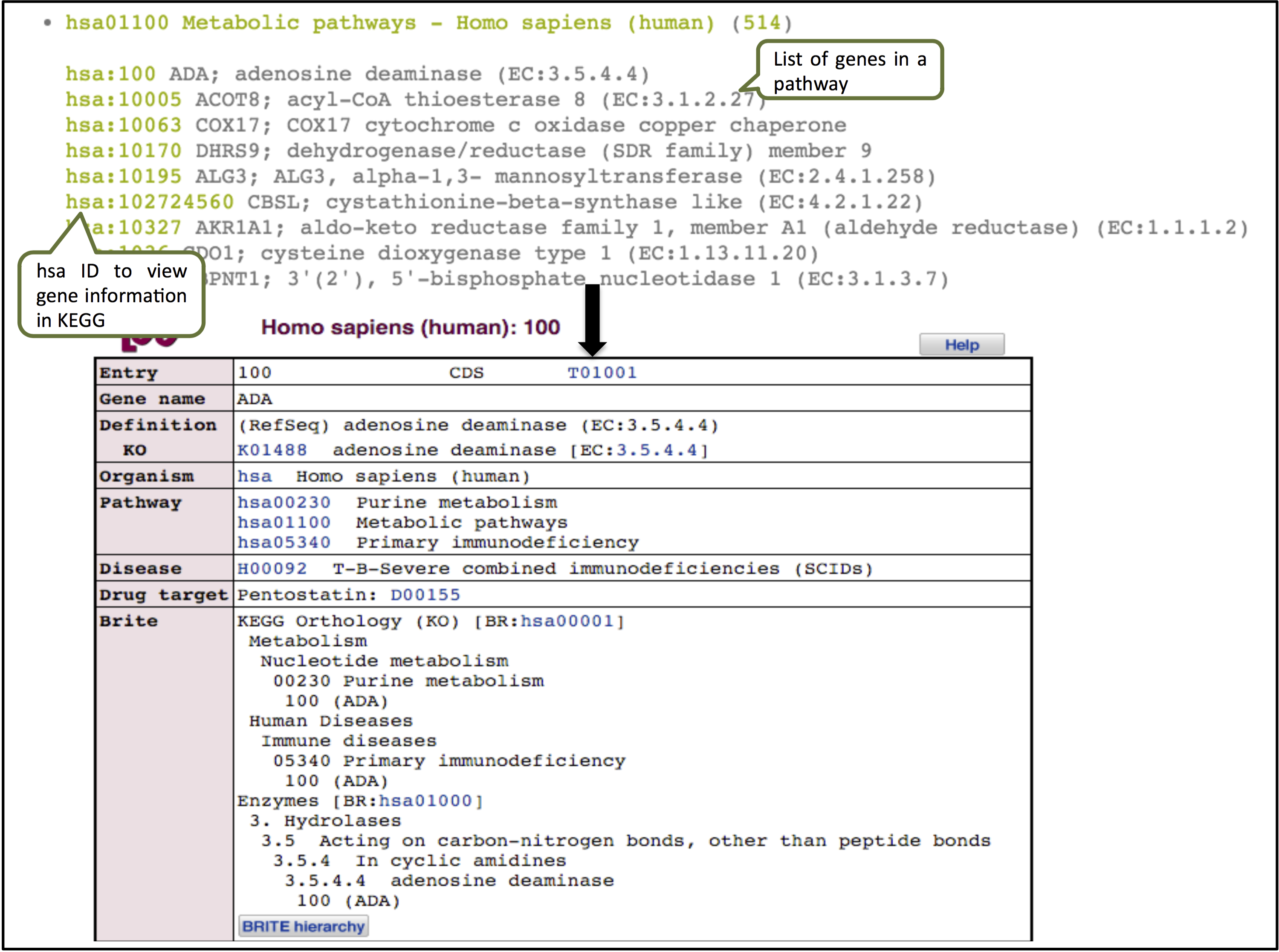
The enrichment tools gives a functional annotation of the genes using KEGG Mapper.
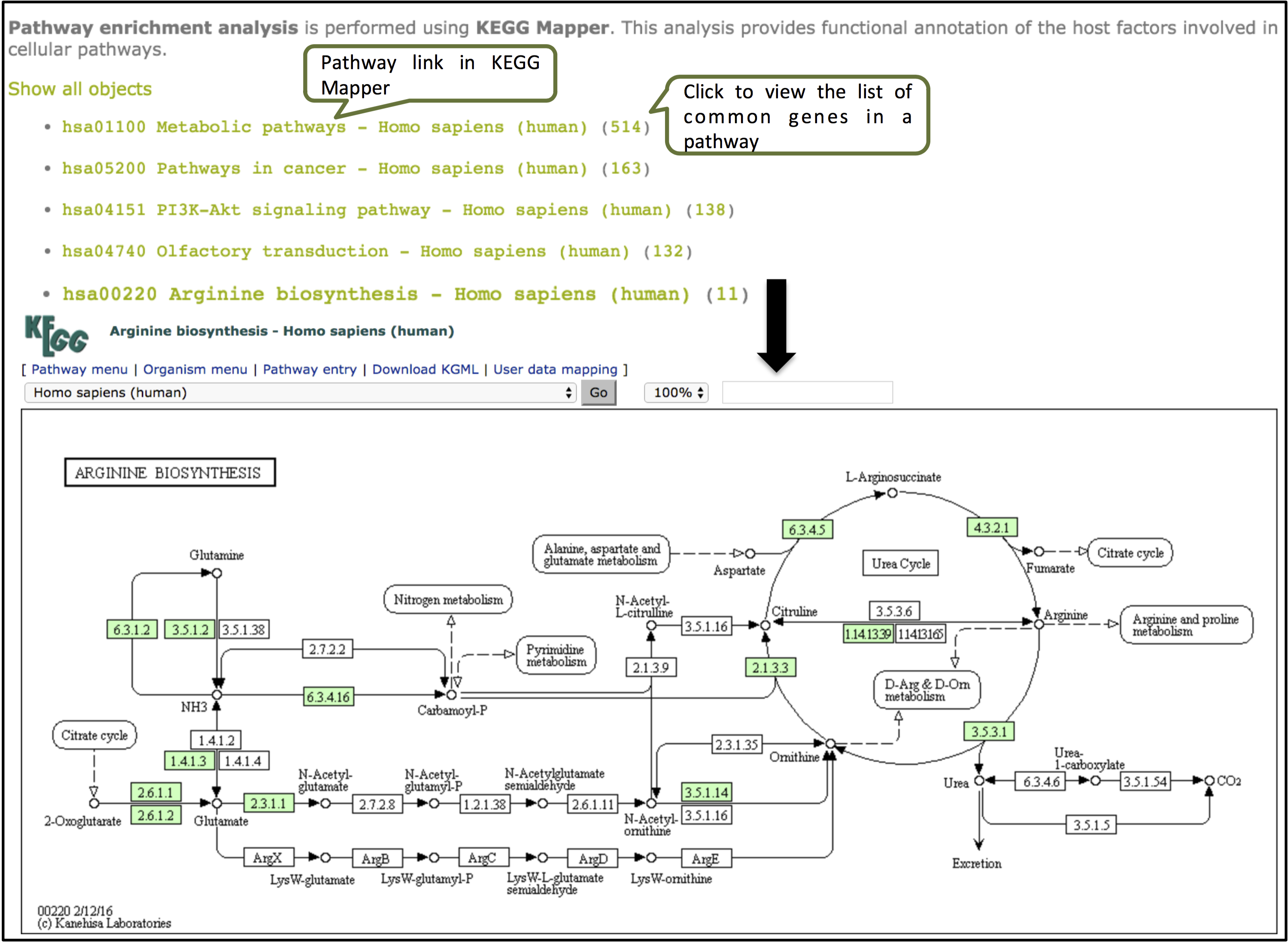
Overlap Analysis
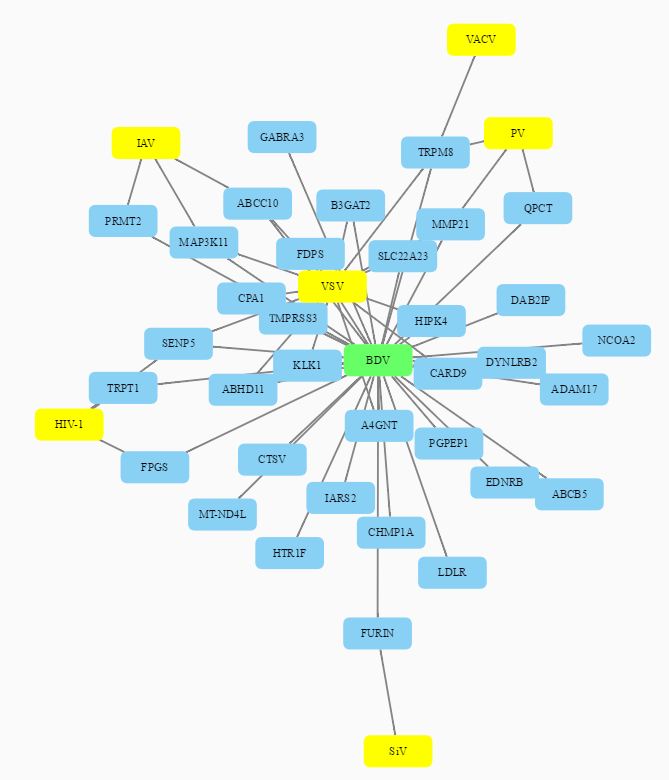
Overlap analysis is used to identify common genes (host factors + restriction factors) between different viruses. User can also find overlapping genes between different virus families and their groups.

Pathogen Interaction
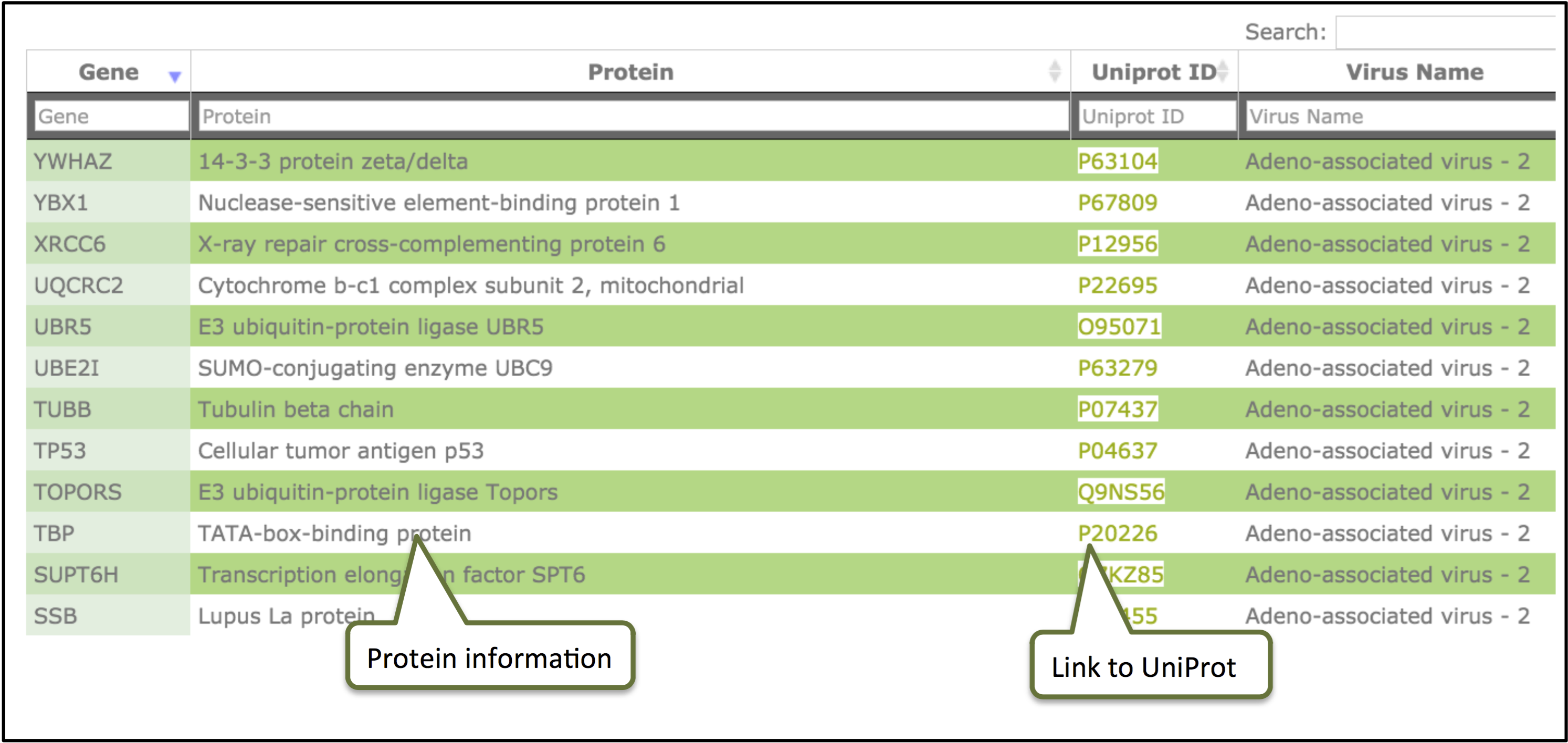
In the pathogen interacting proteins analysis, protein interaction data from VirHostNet and VirusMINT has been provided for further understanding of host-virus biology.
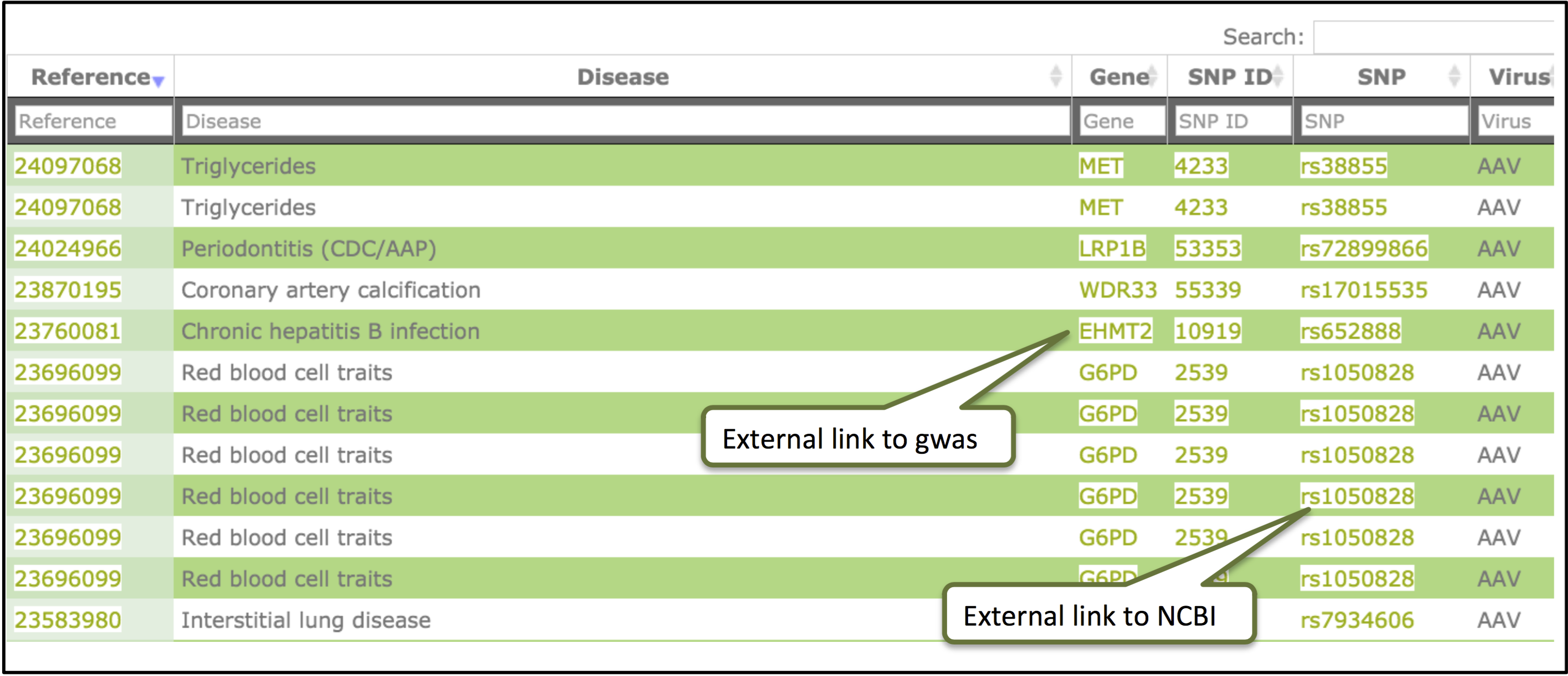
GWAS analysis
In the gwas analysis, user can analyze candidate disease host genes identified from genome wide association studies (NHGRI).