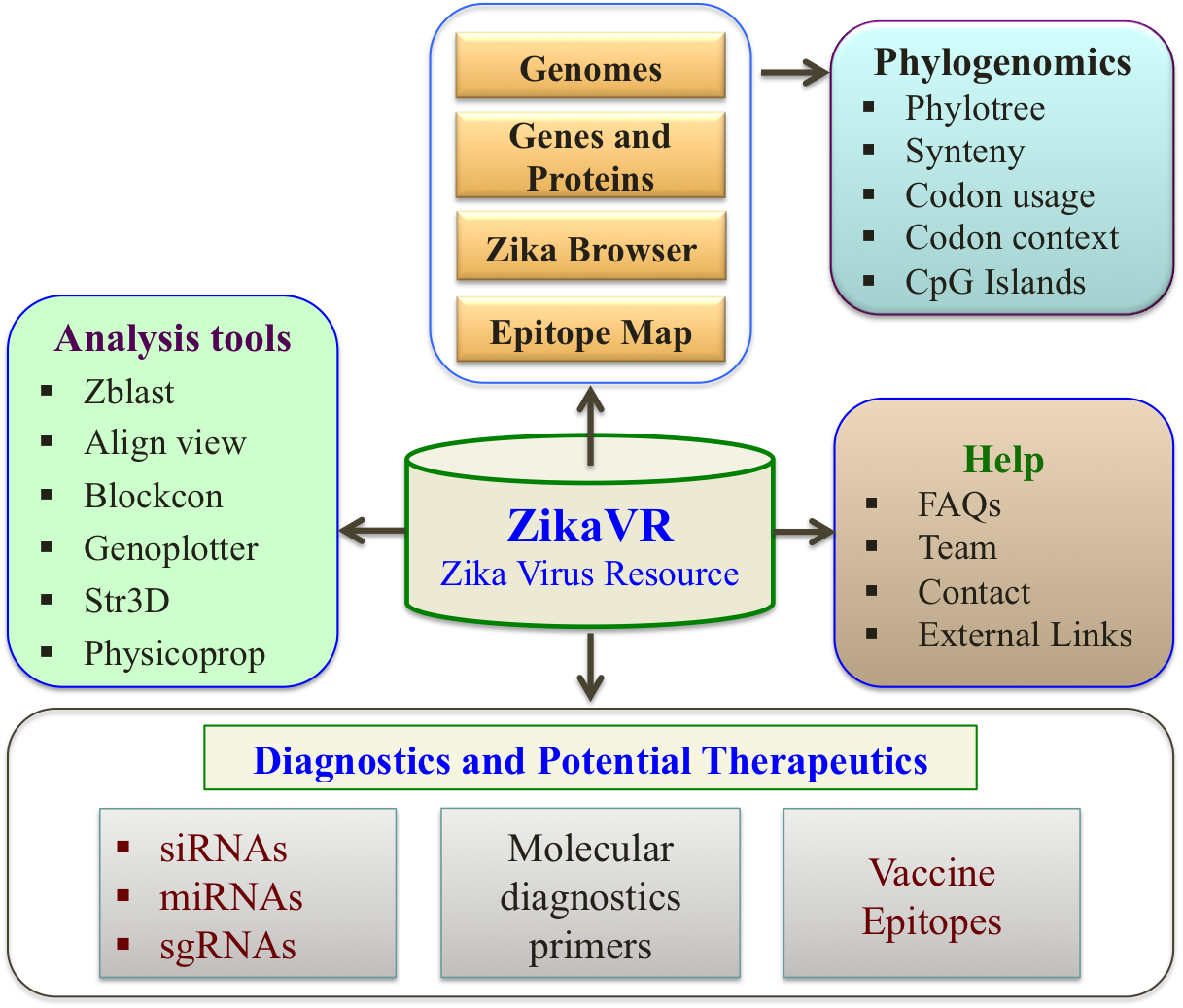
- ZikaVR Overview
- ZikaVR, an integrative multi-omics platform dedicated to the Zika virus genomic and proteomic analysis along with comparative genomics and therapeutic knowledge.
- It is classified in different sections representing individual components i.e. genomic annotation, phylogenomics, therapeutics, comparative genomic analysis and tools.
- It delivers sophisticated analysis such as whole-genome alignments, conservation and variation, CpG islands, GC content, codon usage bias, codon context, synteny and phylogenetic inferences at whole genome; gene and protein level with user-friendly visual environment.
- It also includes graphical Zika genome browser, Zika browser for the collective representation of ZIKV annotation and regulatory information.
- Furthermore, ZikaVR also deals with therapeutically imperative constituents i.e. molecular diagnostics primers, siRNAs, miRNAs, sgRNAs, epitopes, and pathway or network information. An Epitope Map browser is also implemented in resource.
- Detail organization
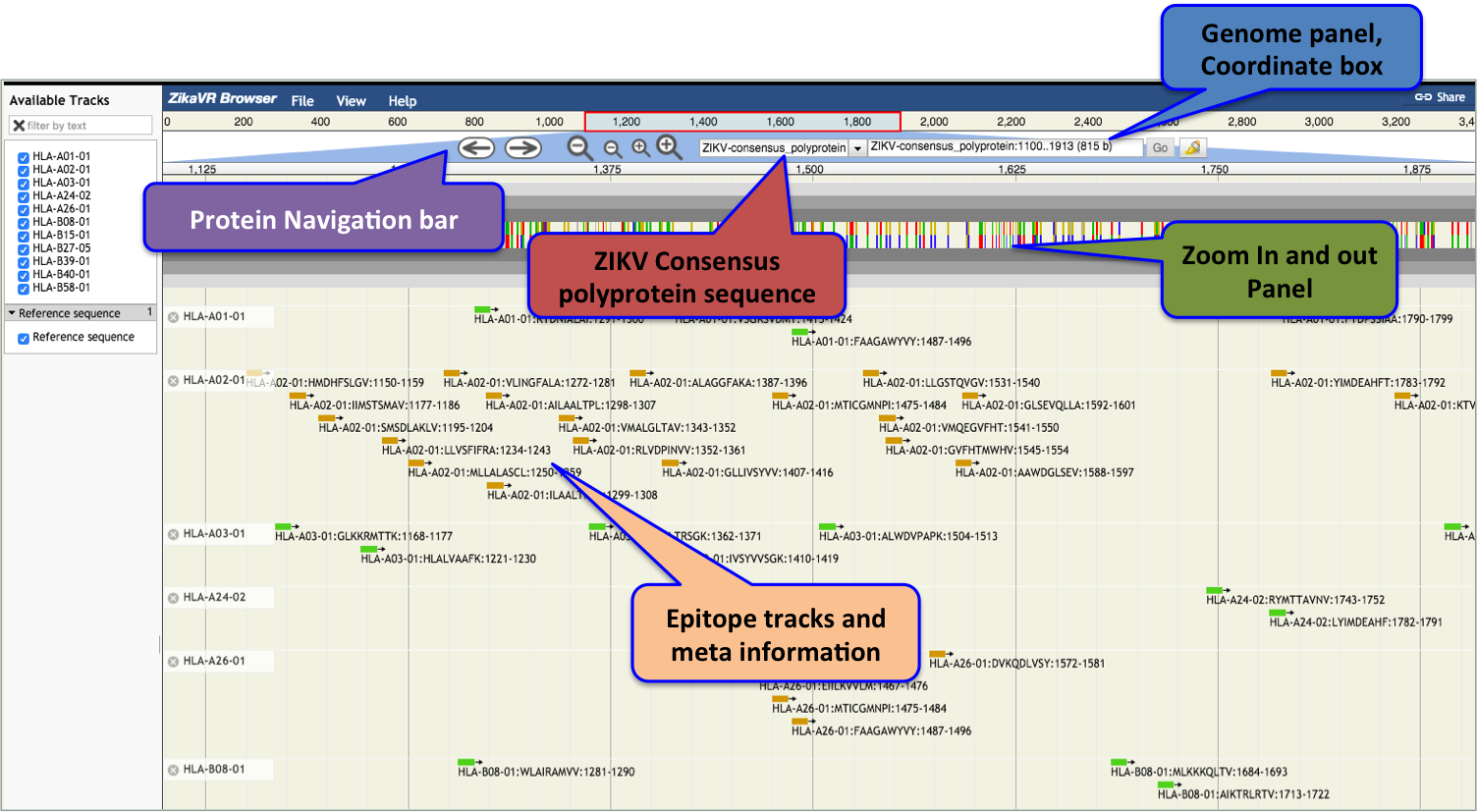
- Zika Browser
- To navigate through the genomes and proteomes, we set up an "Zika genome browser" which facilitates graphical dynamic visualization of annotations.
- Various tracks includes Zika genome, proteome, regions, and regulatory sites
- The upper panel shows the positional scale (ruler) to navigate through genomes/proteome along with reference sequence. Distinct annotation features were shown in separate color blocks.

- Epitope Map
- This is an interactive map of epitopes. In Epitope map, data is classified according to the HLA types.
- It contains information such as Epitope id, start and end coordinates with gene region, epitope length, epitope sequence, and HLA allele type. Epitopes are mapped on the consensus viral polyprotein sequences. User can map potentially identified novel or predicted epitope sequence using this tool.
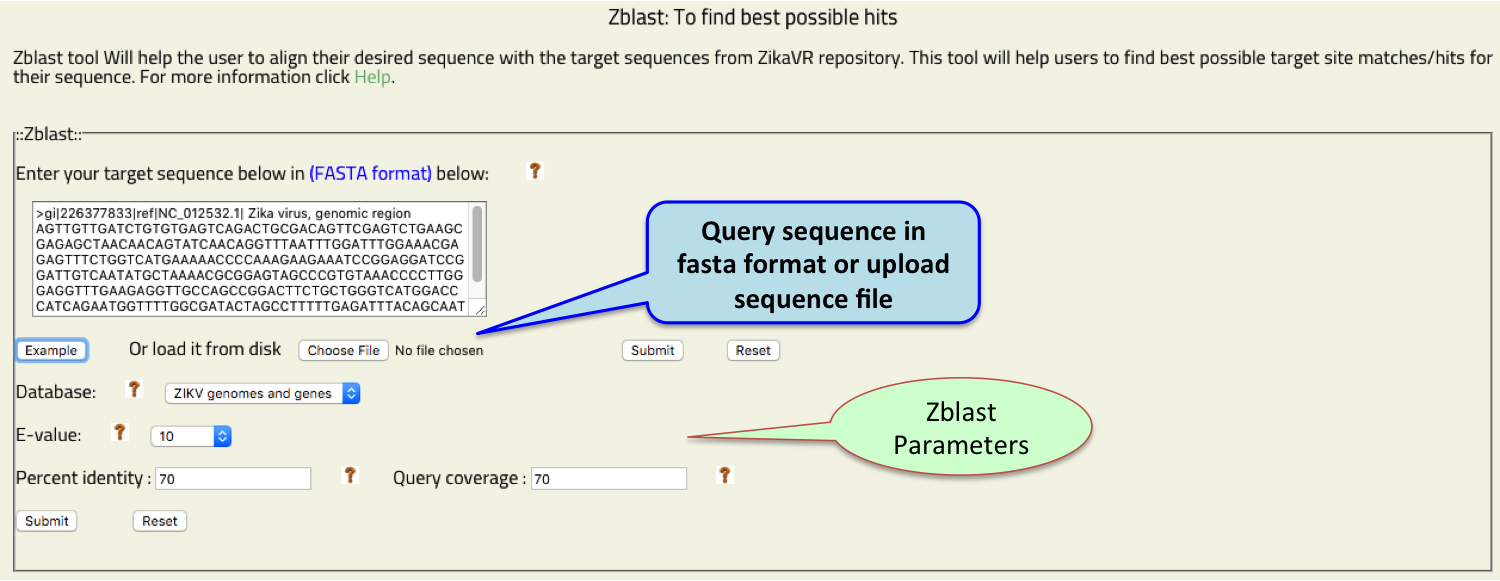
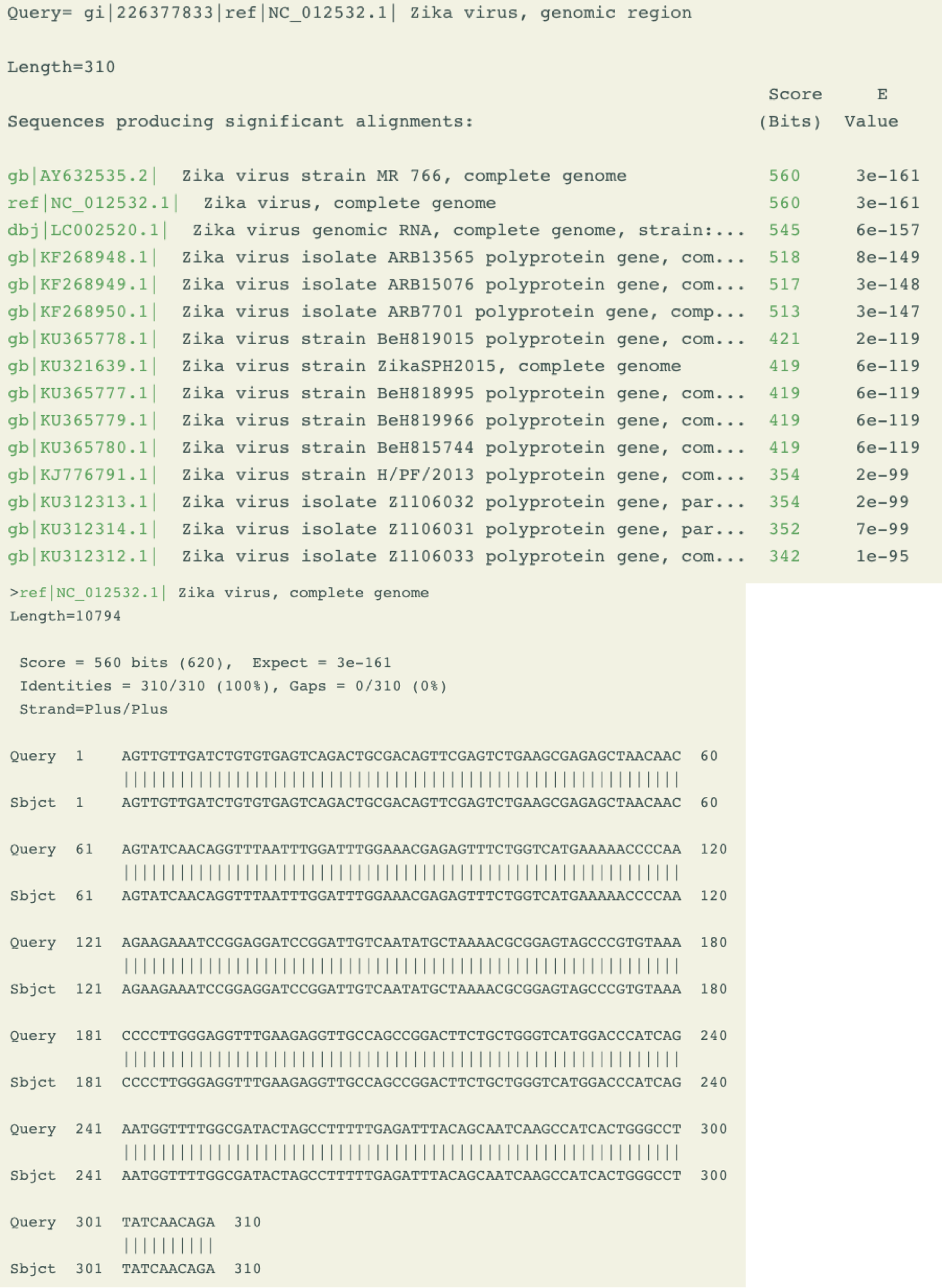
- Zblast
- Zblast:: To find similarity and align query sequence to the Zika genomes and genes.
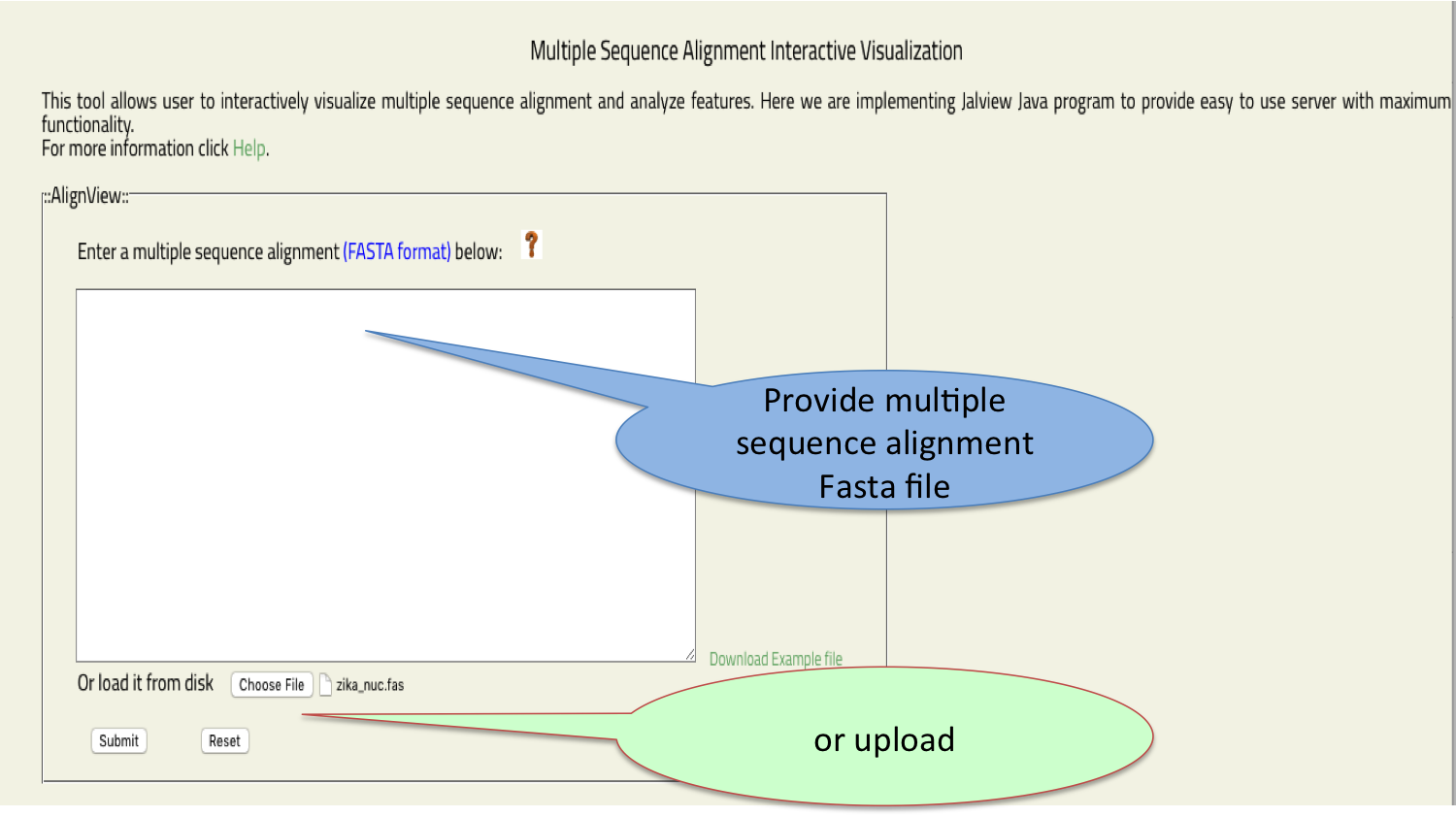
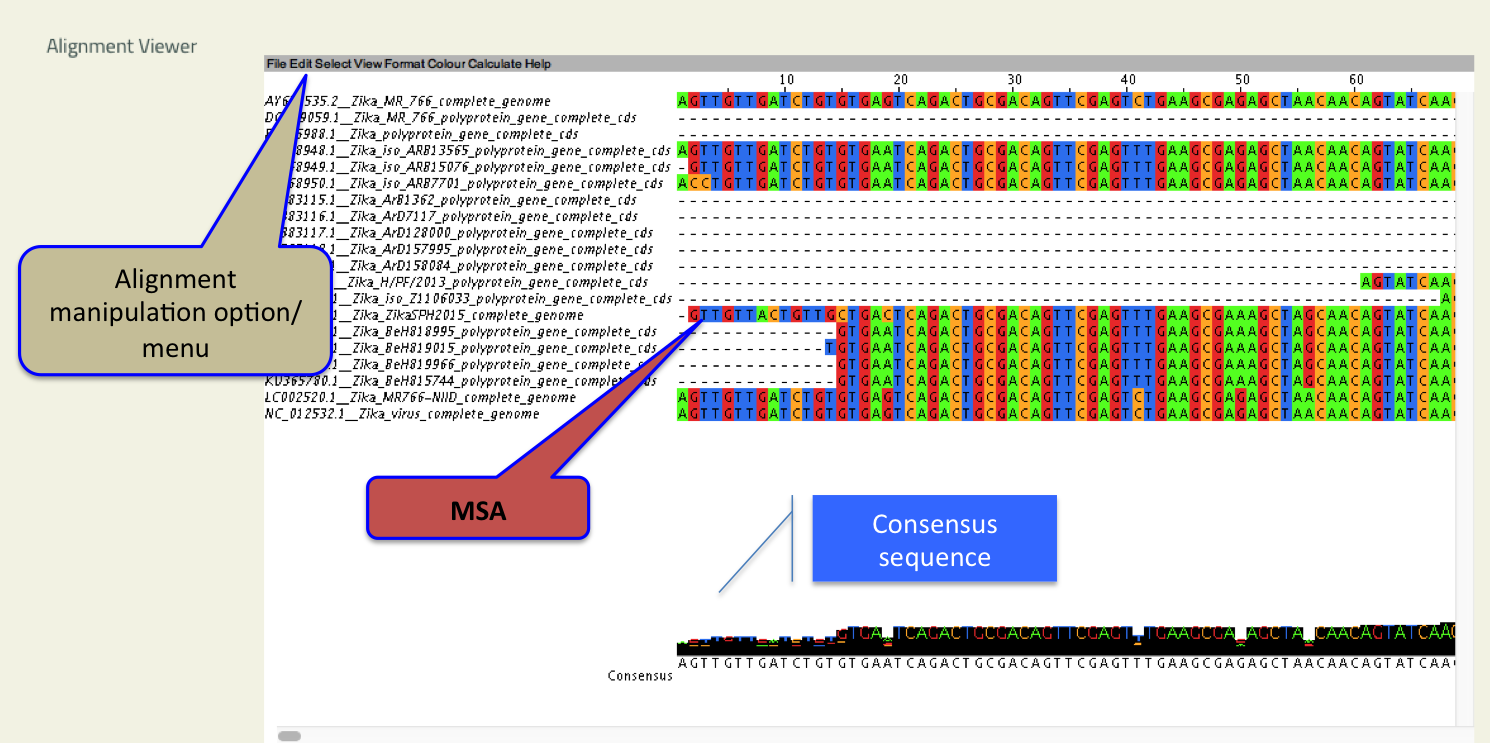
- Align View
- This tool allows user to interactively visualize multiple sequence alignment and analyze features.
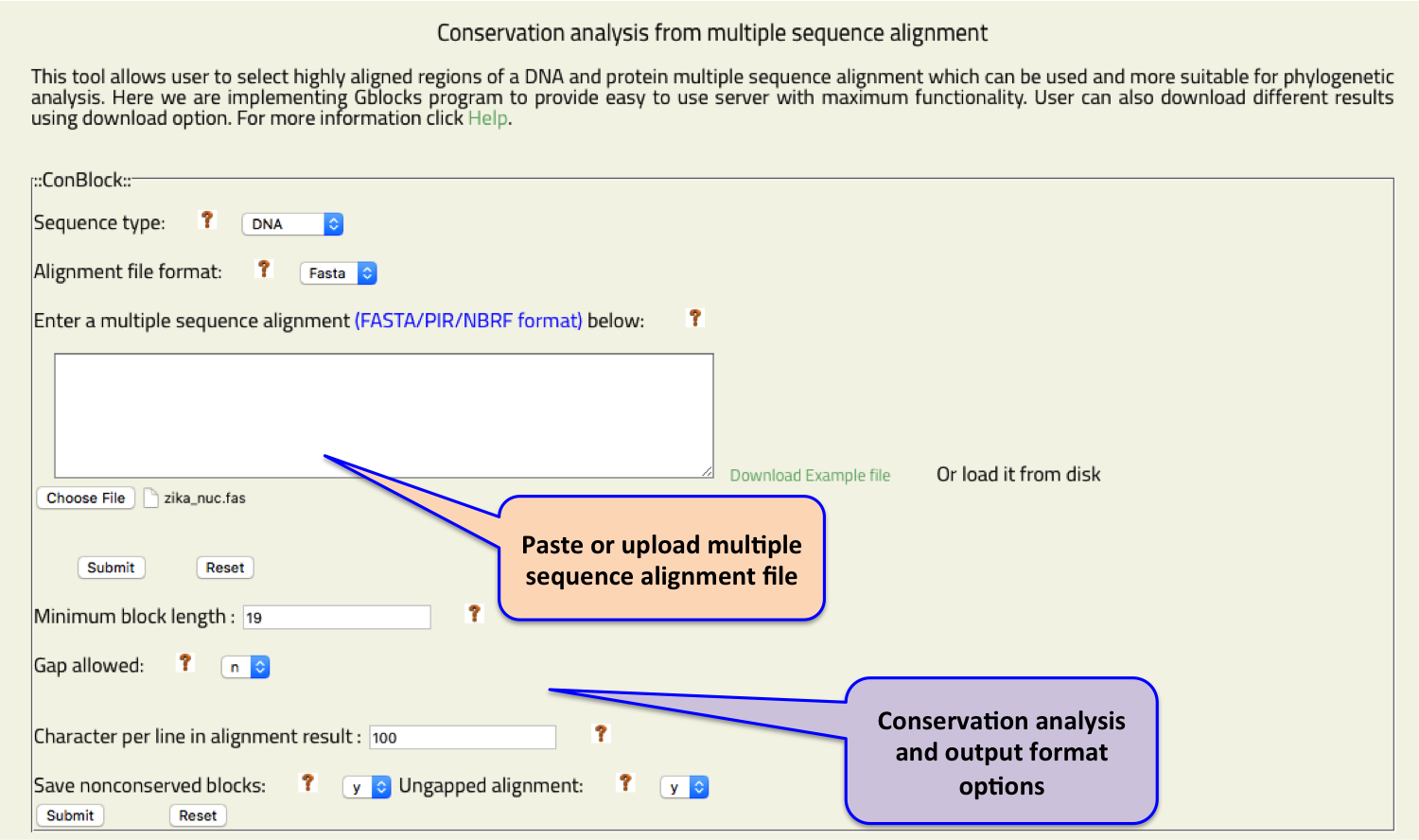
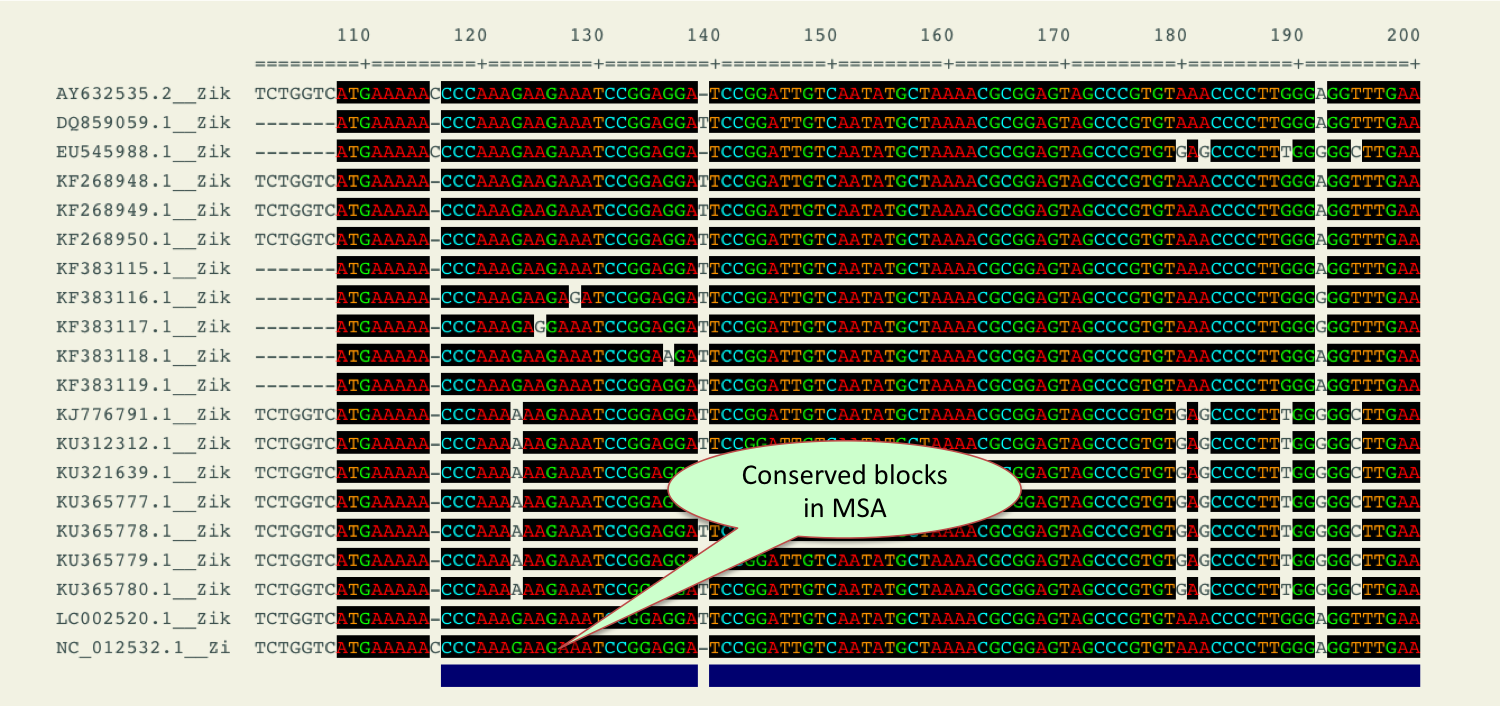
- Blockcon
- This tool allows user to select highly aligned regions of a DNA and protein multiple sequence alignment which can be used and more suitable for phylogenetic analysis.
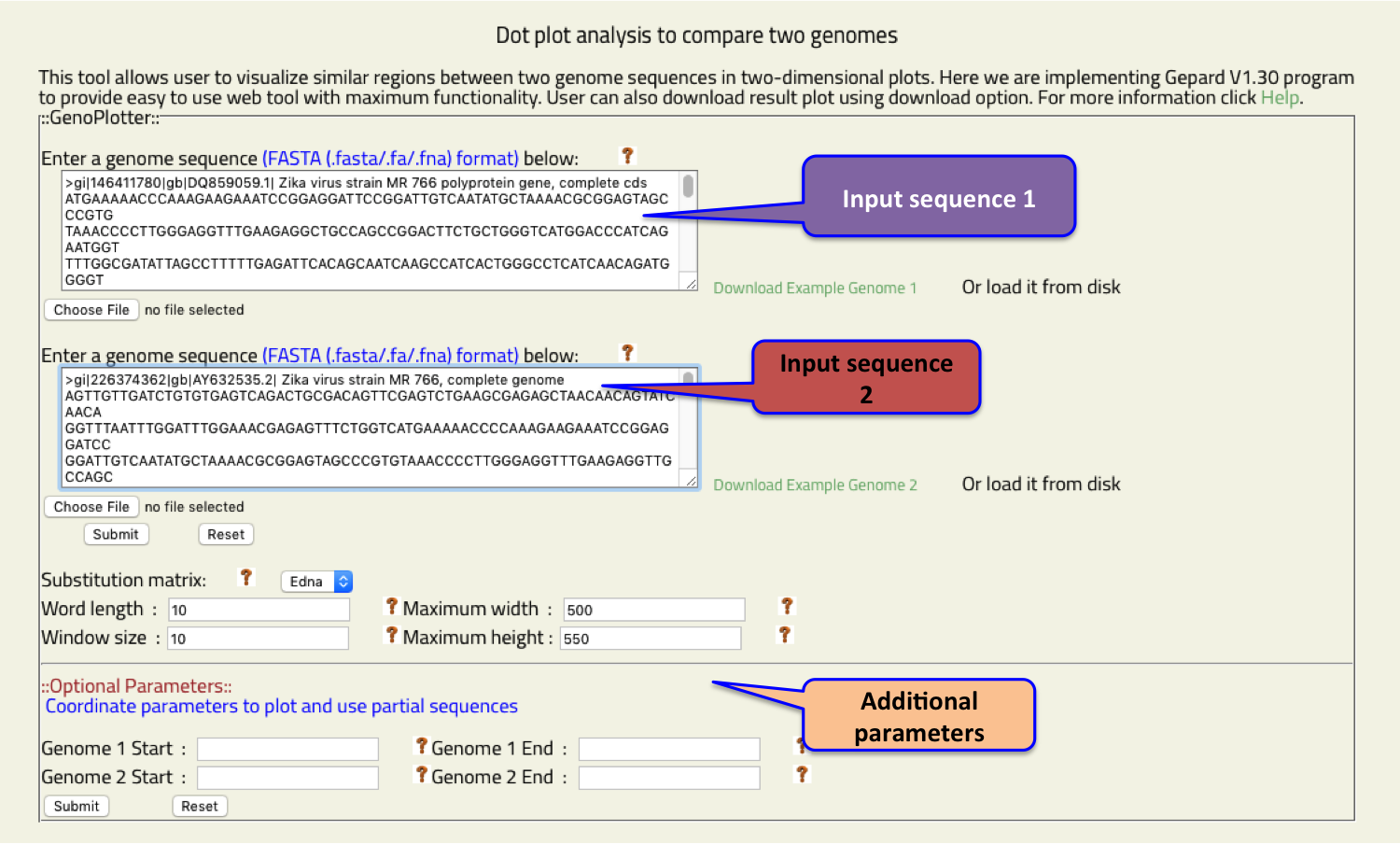
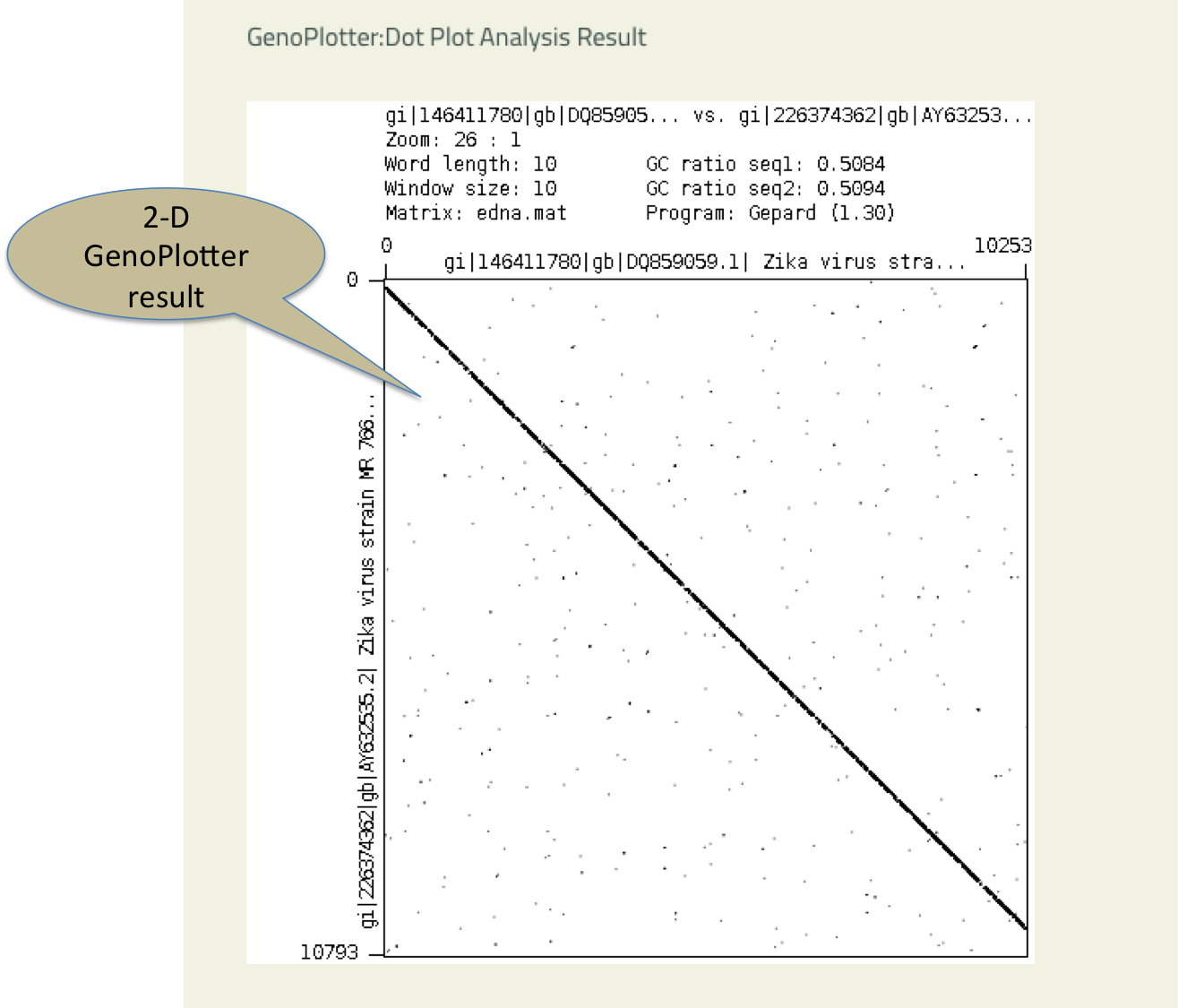
- Genoplotter
- This Dot plot analysis tool allows user to compare two genomes.
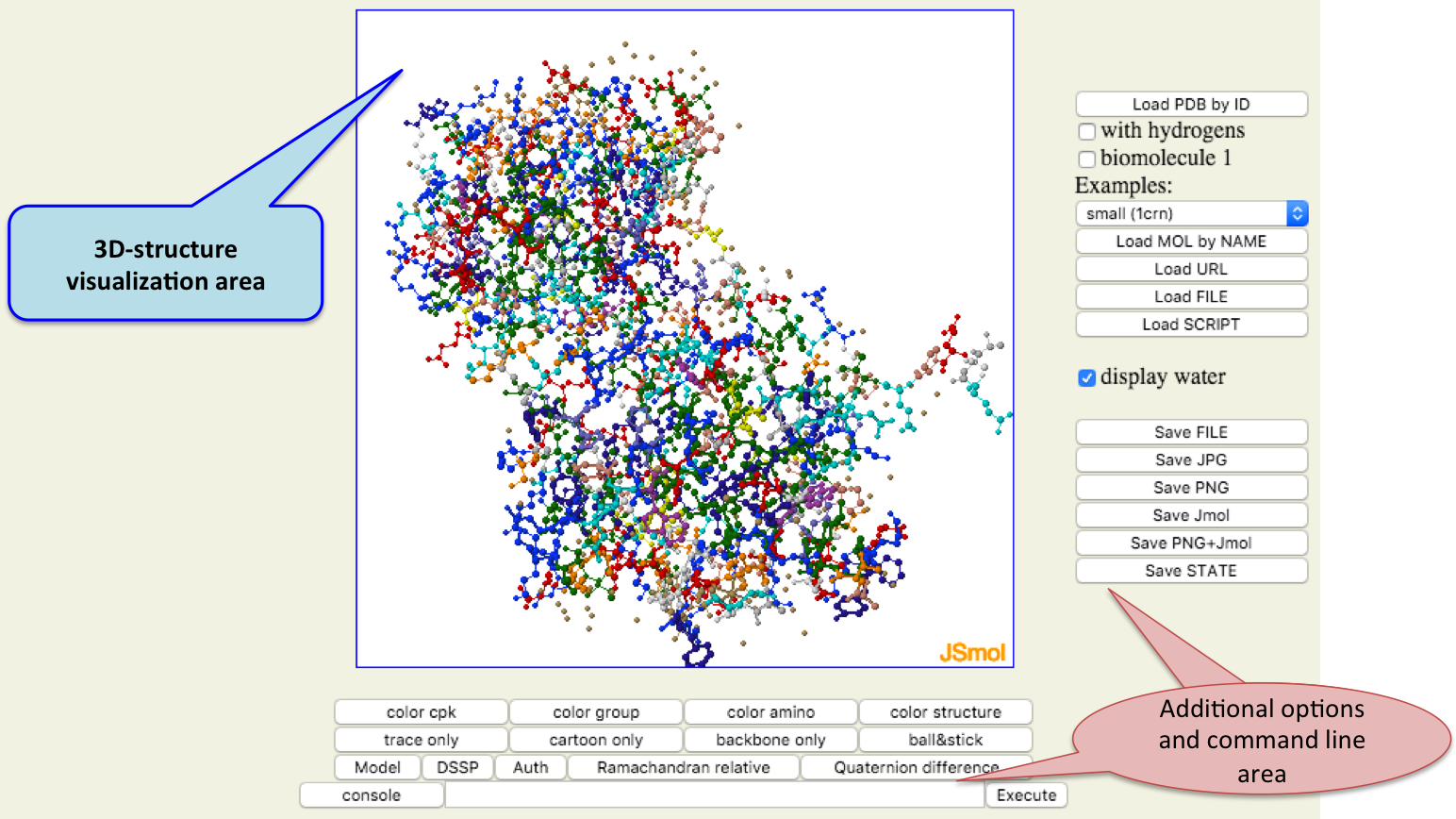
- Str3D
- Str3D:: 3D-structure visualization of proteins and molecules.
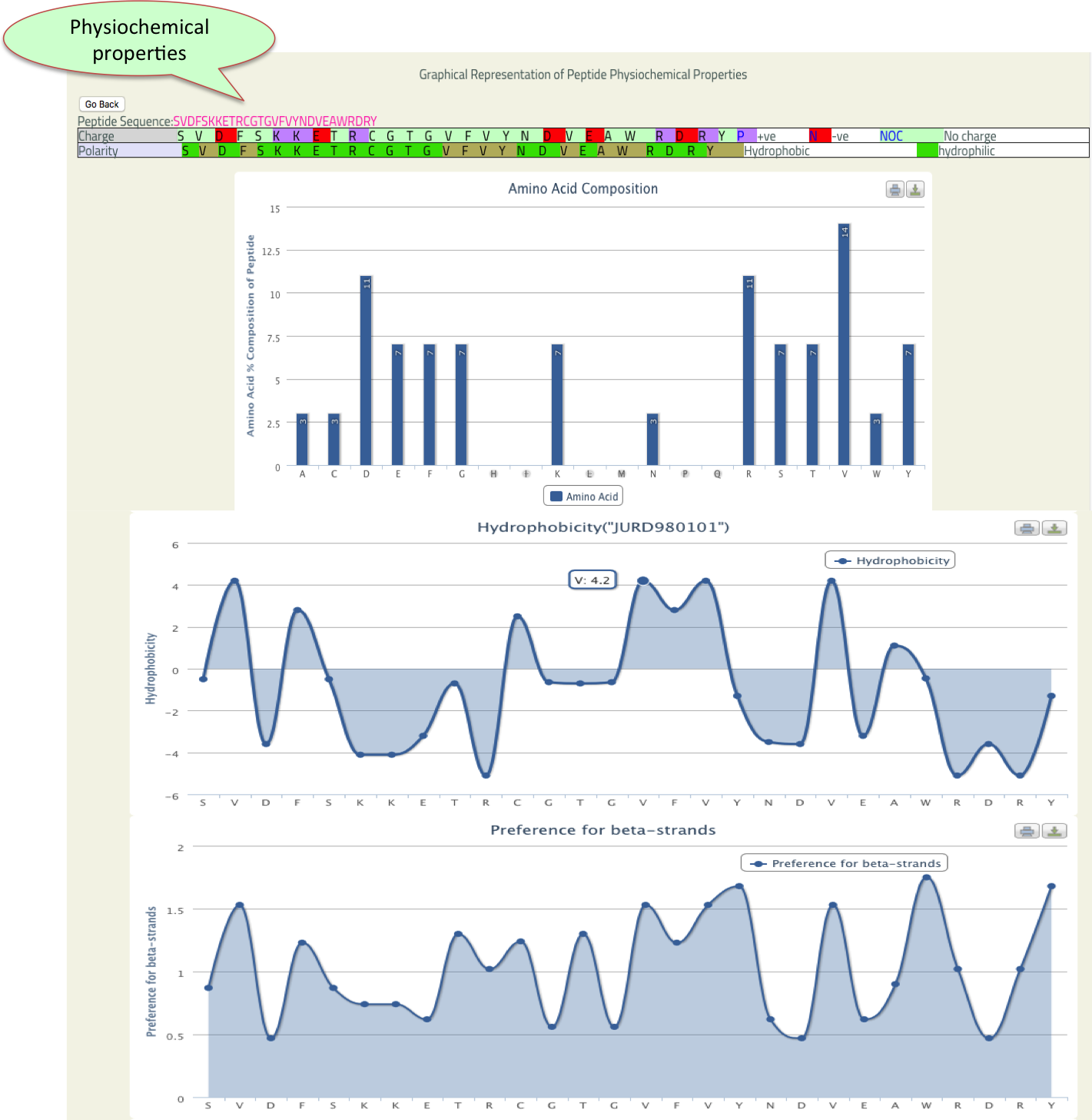
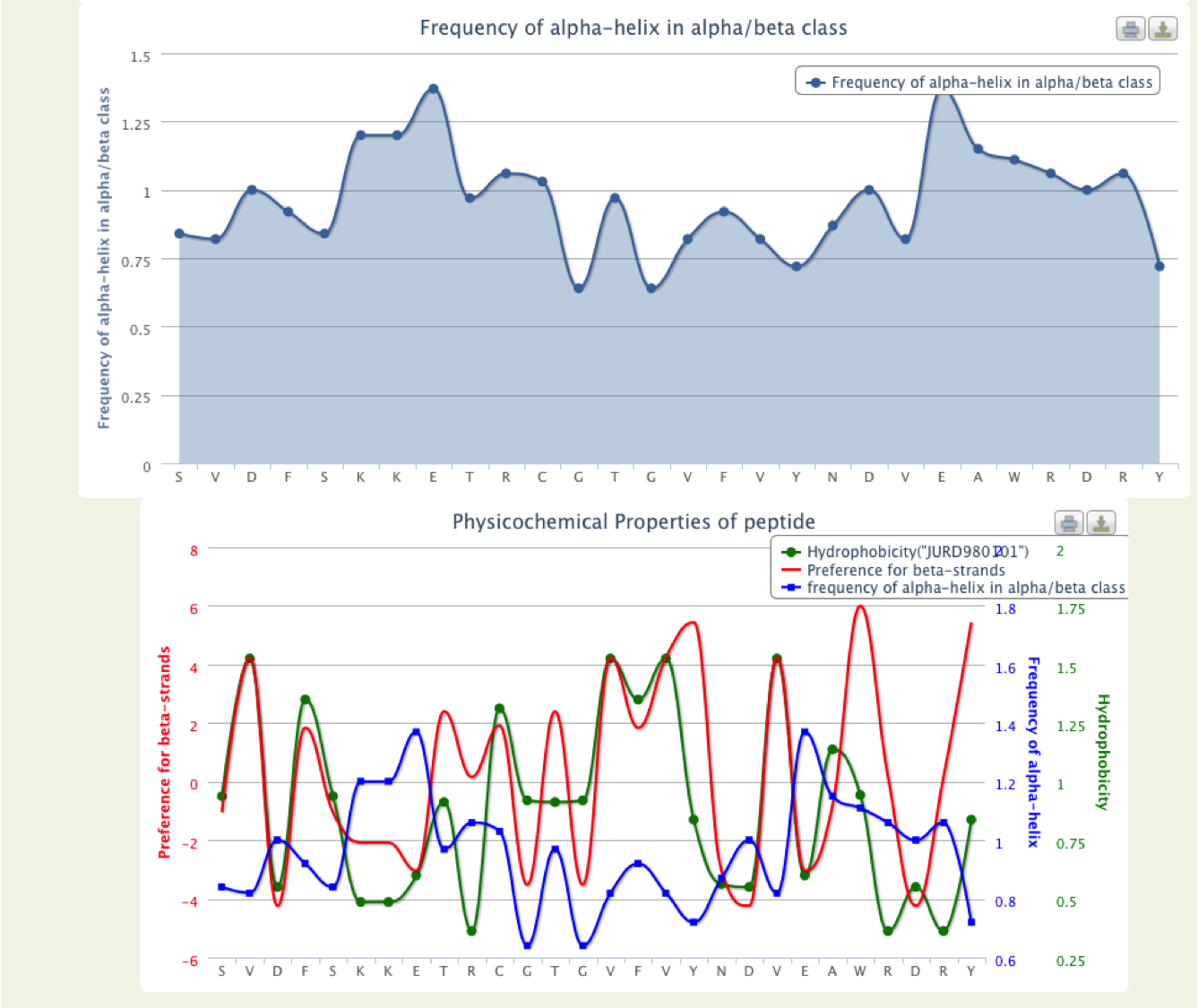
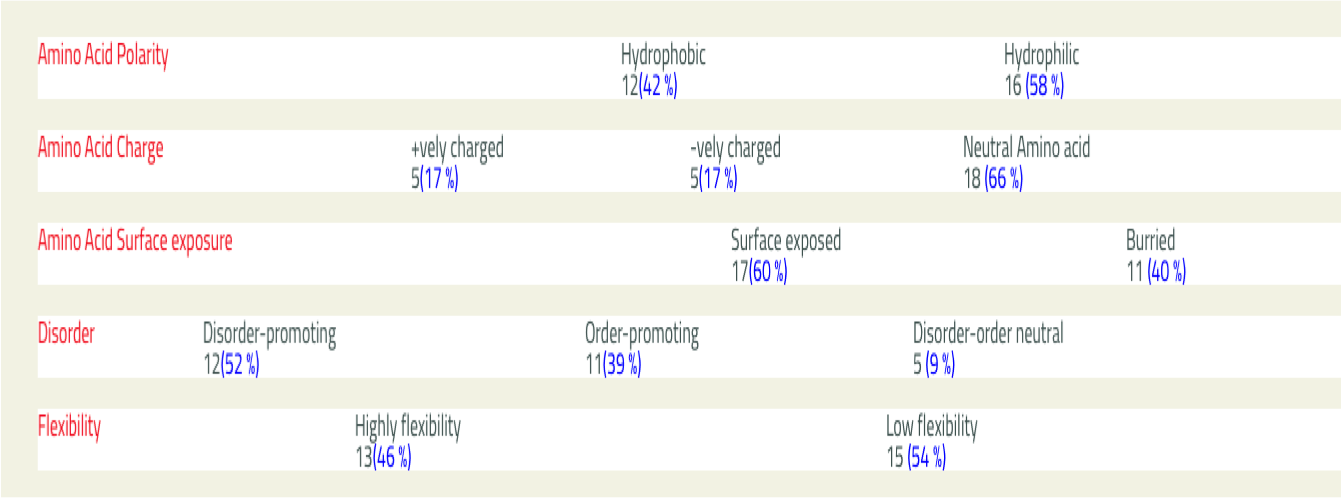
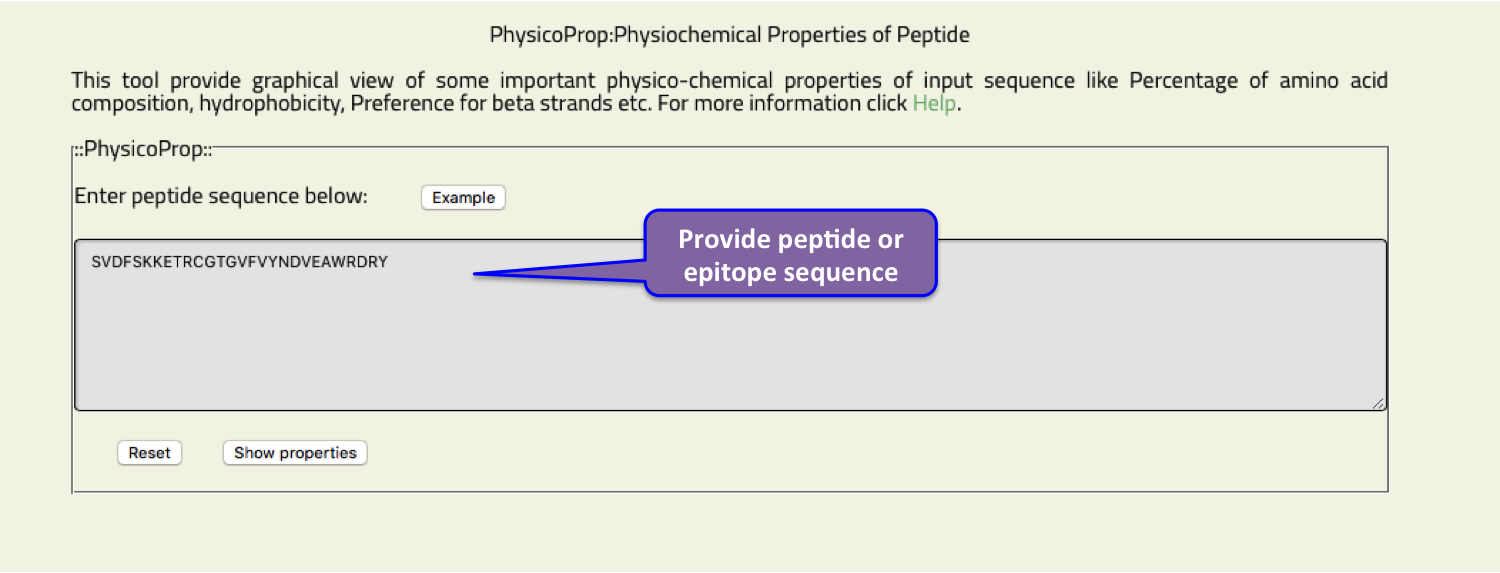
- PhysicoProp
- This tool provide graphical view of some important physico-chemical properties of input sequence like Percentage of amino acid composition, hydrophobicity, Preference for beta strands etc.
- Therapeutics
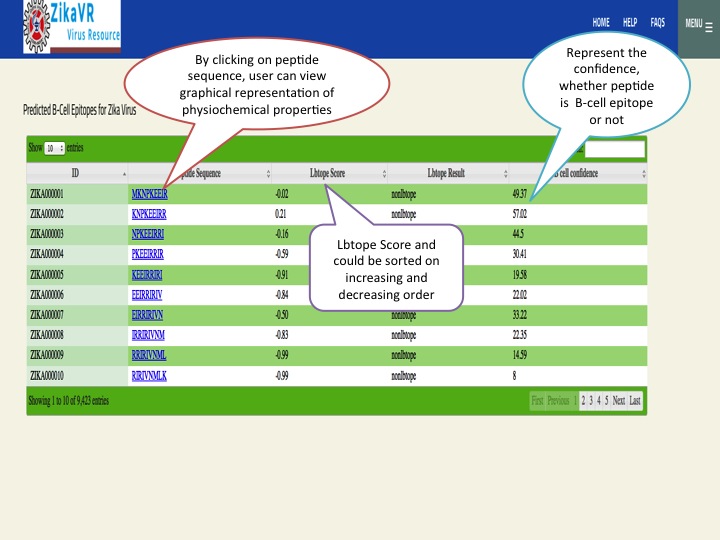
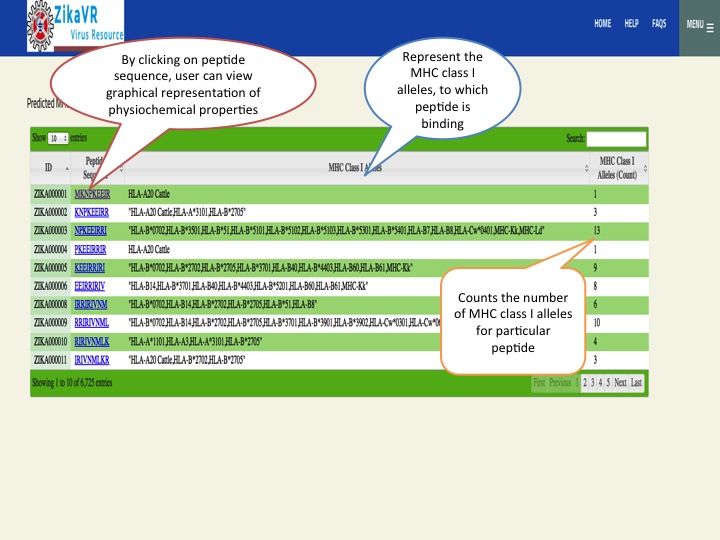
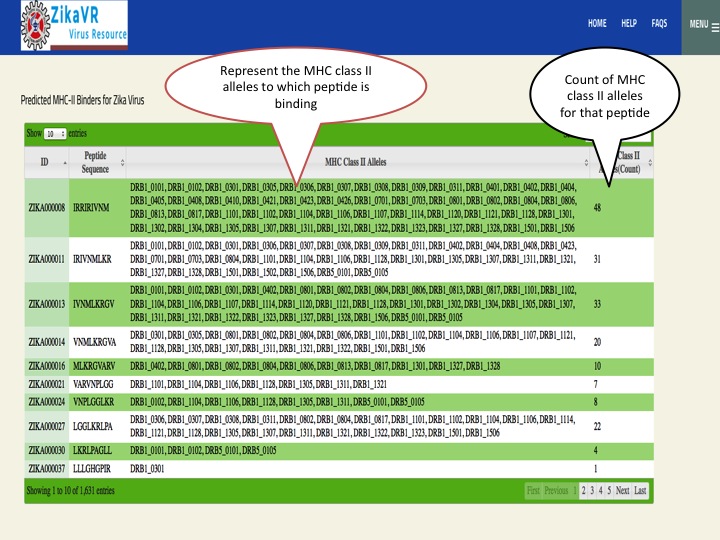
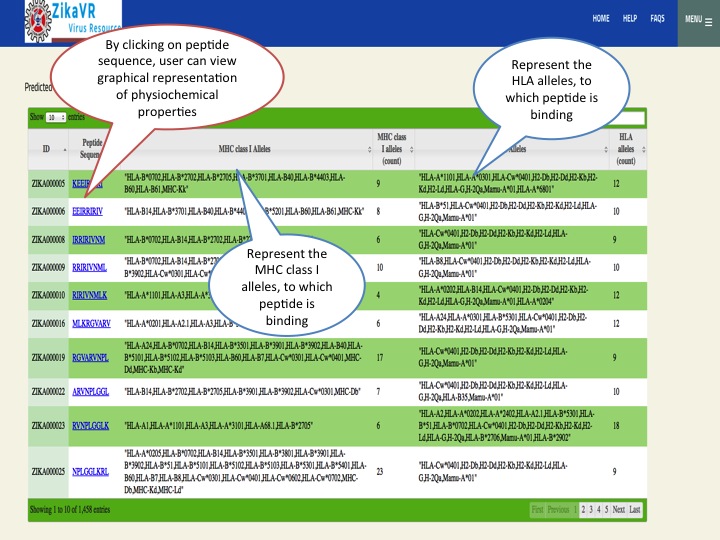
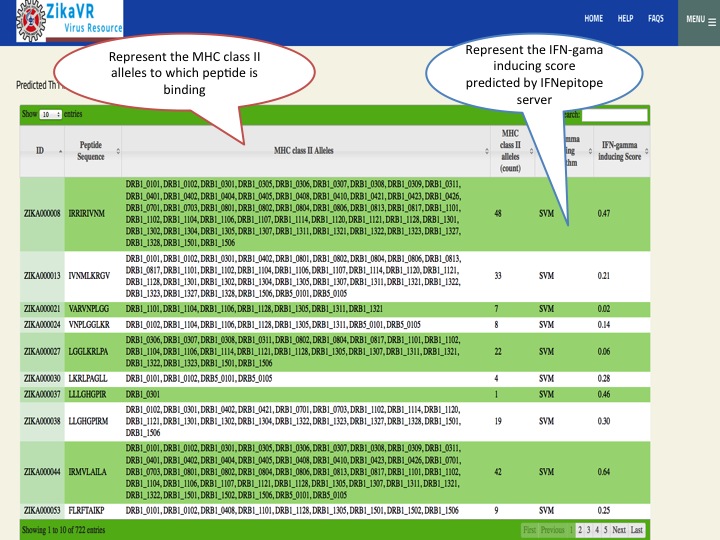
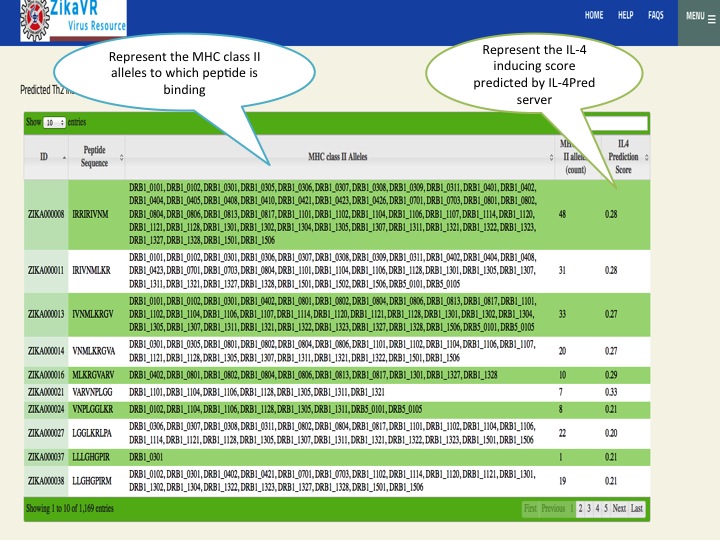
- We have predicted potential subunit vaccine candidates for Zika virus using various compuational algorithm. We used a pipeline for predicting the epitopes.This pipeline have the package for predicting linear B cell epitopes using LBtope, promiscous binder of MHC class I (Propred1) and MHC class II (Propred), CTL epitope (CTLpred), interferon-gamma inducing peptides (IFNepitope) and Interleukine 4 inducing peptides (IL4pred). The peptides can sorted at any header. In this section, we have summarized all the peptides that were able to induce diverse arms of immune response.
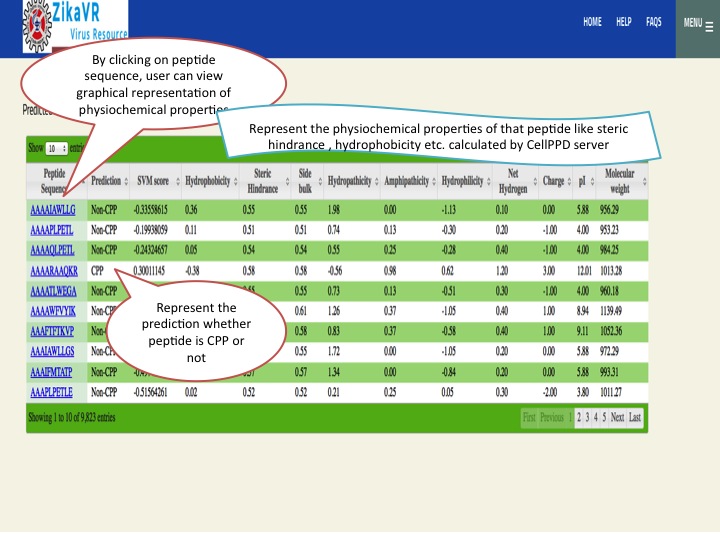
- We have predicted the cell penetrating peptides of zika virus protein using CellPPD server. It predicts potential cell penetraing peptide and calculate physiochemical properties of that peptide like Hydrophobicity, Steric hinderance, Side bulk, Hydropathicity, Amphipathicity, Hydrophilicity, Net Hydrogen Charge, pI and Molecular weight.
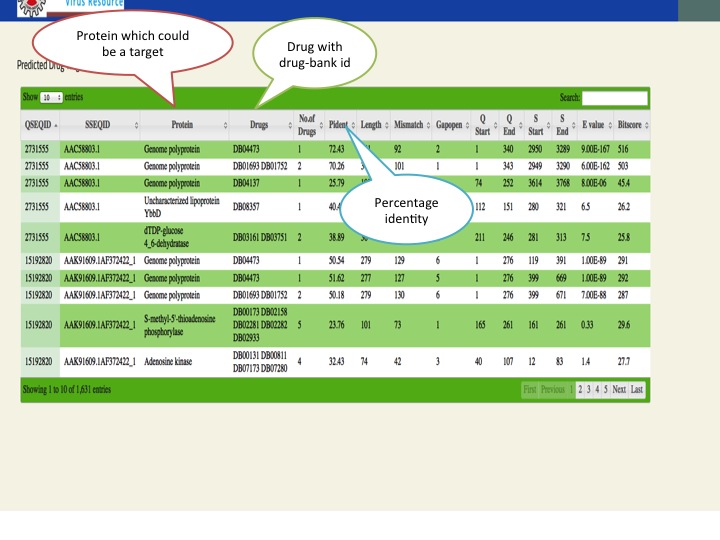
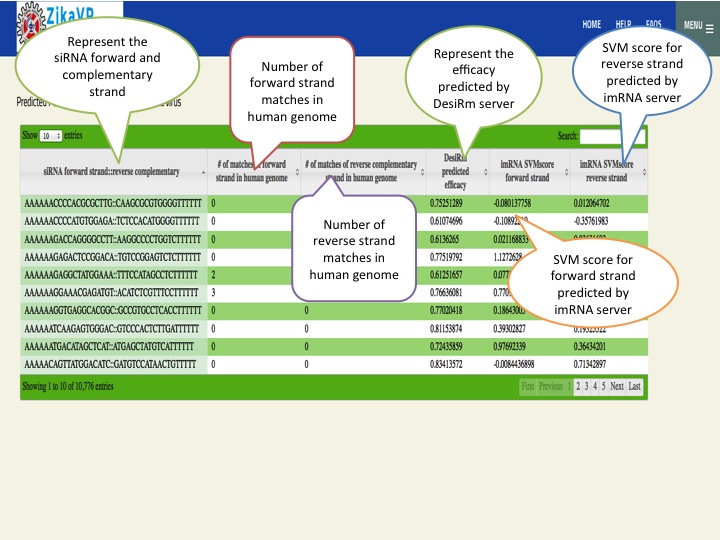
- We have mapped all the proteins of zika virus on drug bank to screen potential similar drug targets. Similarly we have also predicted siRNA suppressor for zika virus as well as immunomodulatory RNA using DesiRm and imRNA server.

Input:
Output:

Input:
Output:
Input:
Output:
Input:
Output:
Input:
Output: