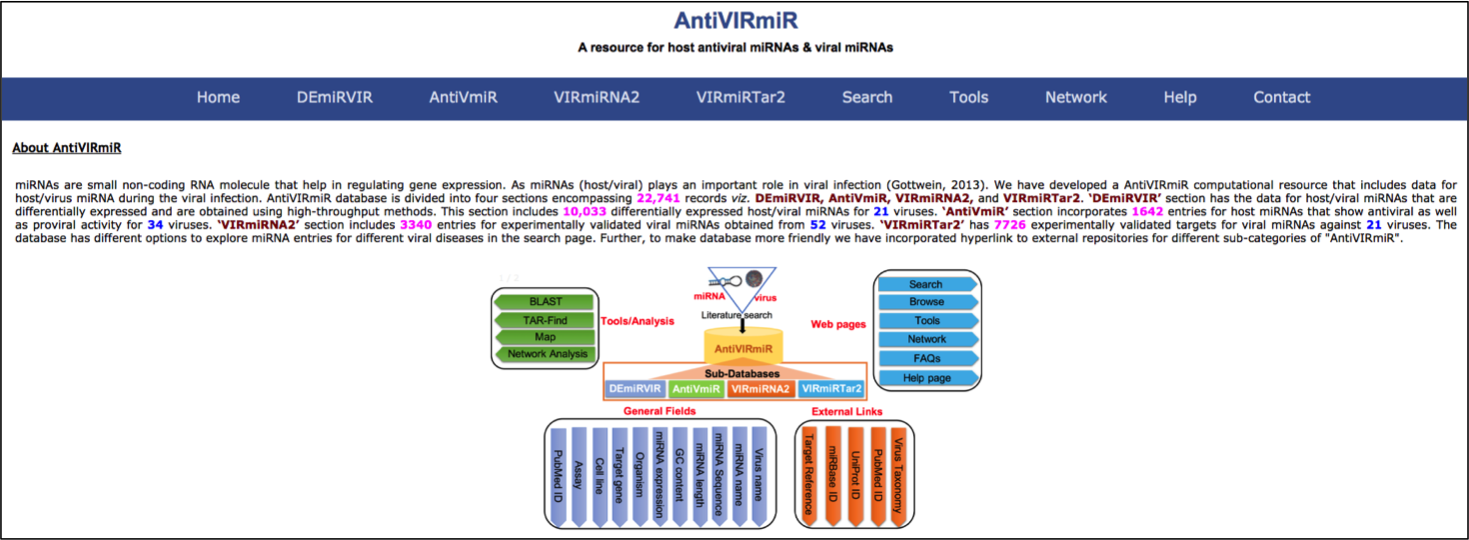
AntiVIRmiR is a computational resource that includes data for host/virus miRNA during the viral infection. AntiVIRmiR resource encompassing 22,741 records is divided into four sections viz. VIRmiRNA2, VIRmiRTar2, AntiVmiR and DEmiRVIR. In ‘DEmiRVIR’ section, we have collected the data for host/viral miRNAs that are differentially expressed and are obtained using high-throughput methods. This section includes 10,033 differentially expressed host/viral miRNAs for 21 viruses.‘AntiVmiR’ section incorporates 1642 entries for host miRNAs that show antiviral as well as proviral activity for 34 viruses.‘VIRmiRNA2’ section includes 3340 entries for experimentally validated viral miRNAs obtained from 56 viruses. ‘VIRmiRTar2’ has 7726 experimentally validated targets for viral miRNAs against 21 viruses.

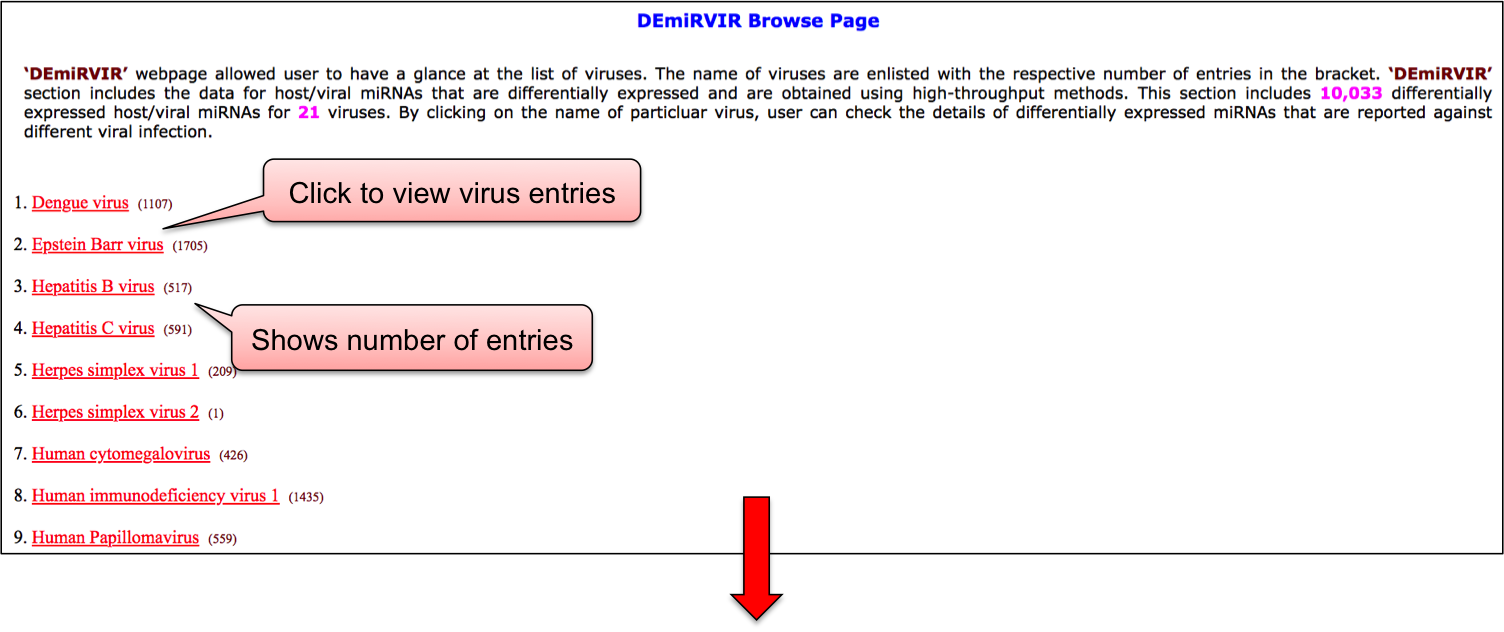
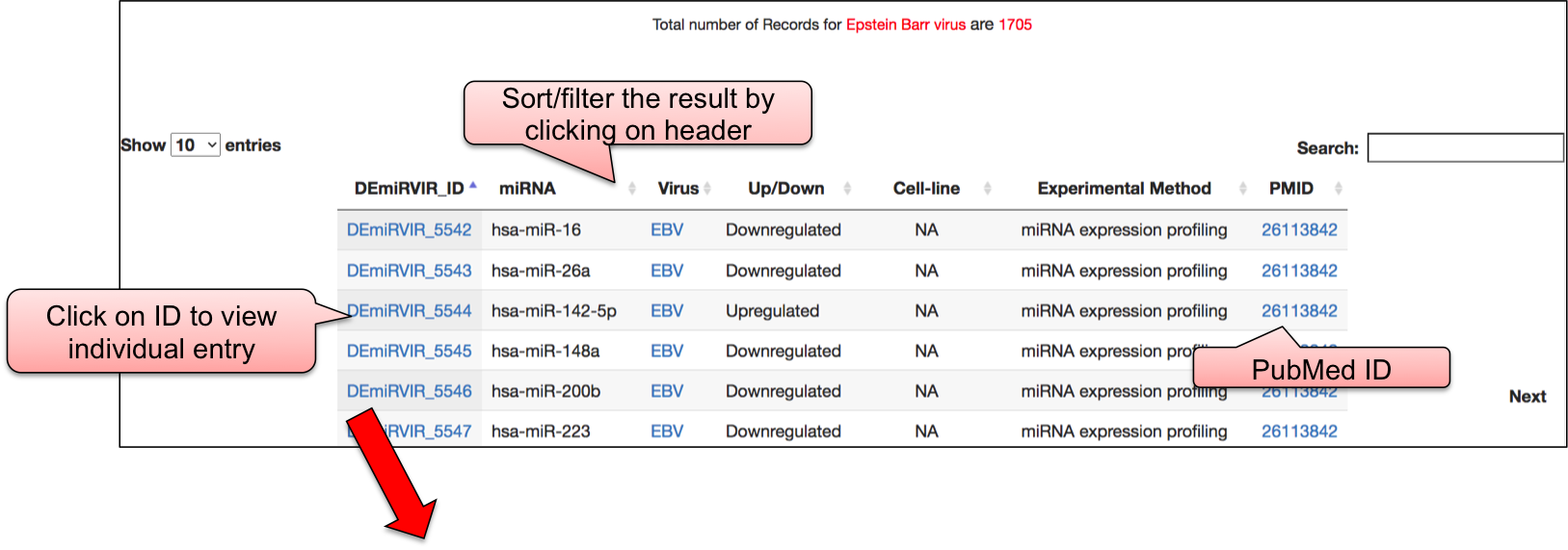
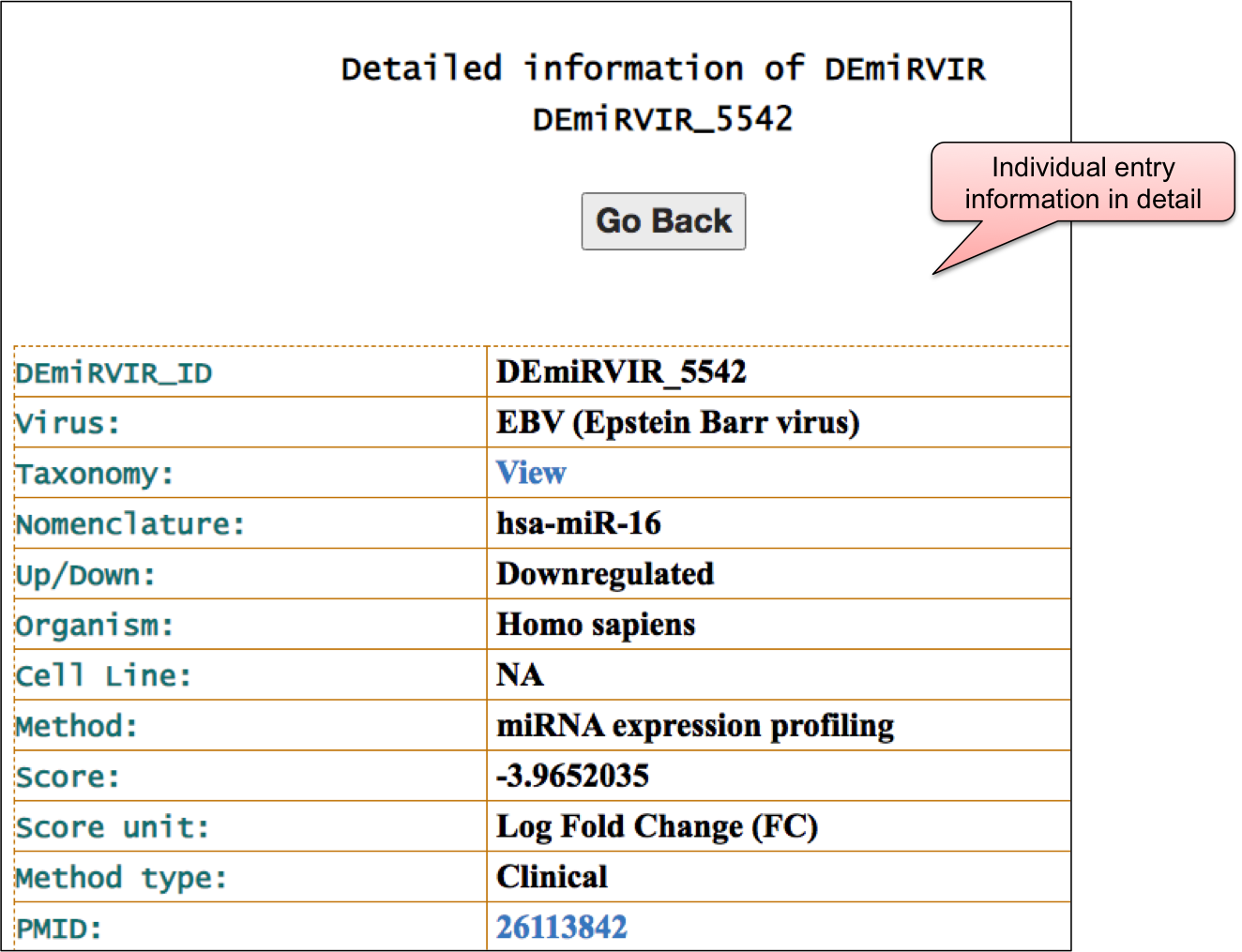
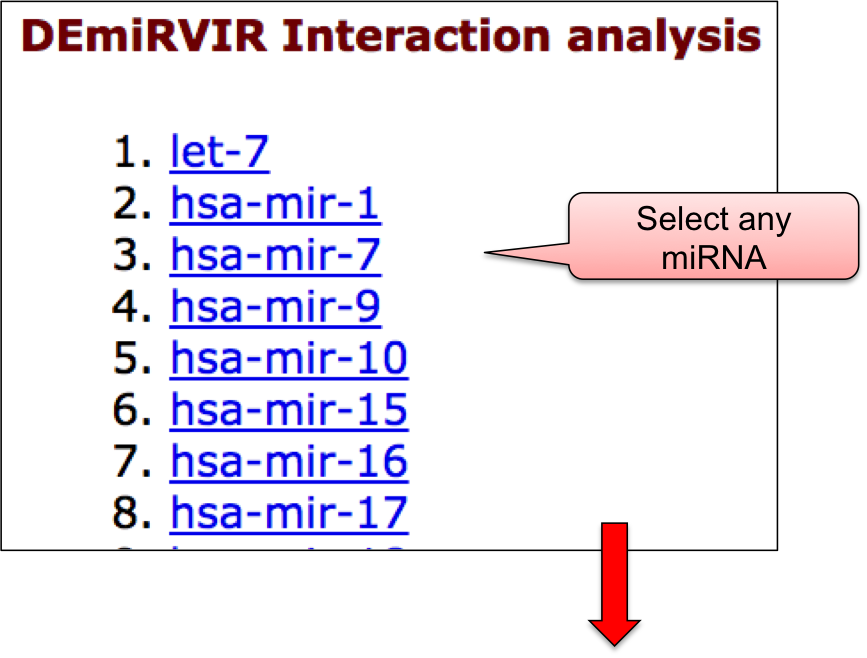
‘DEmiRVIR’ webpage allowed user to have a glance at the list of viruses. The name of viruses are enlisted with the respective number of entries in the bracket. ‘DEmiRVIR’ section includes the data for host/viral miRNAs that are differentially expressed and are obtained using high-throughput methods. This section includes 10,033 differentially expressed host/viral miRNAs for 21 viruses. By clicking on the name of particluar virus, user can check the details of differentially expressed miRNAs that are reported against different viral infection.
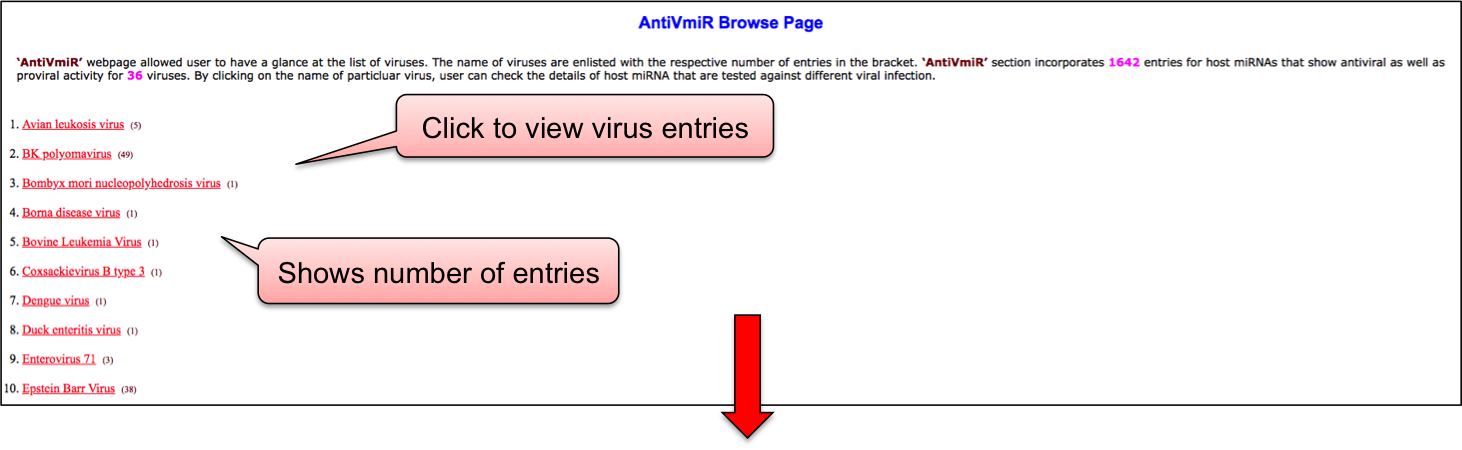
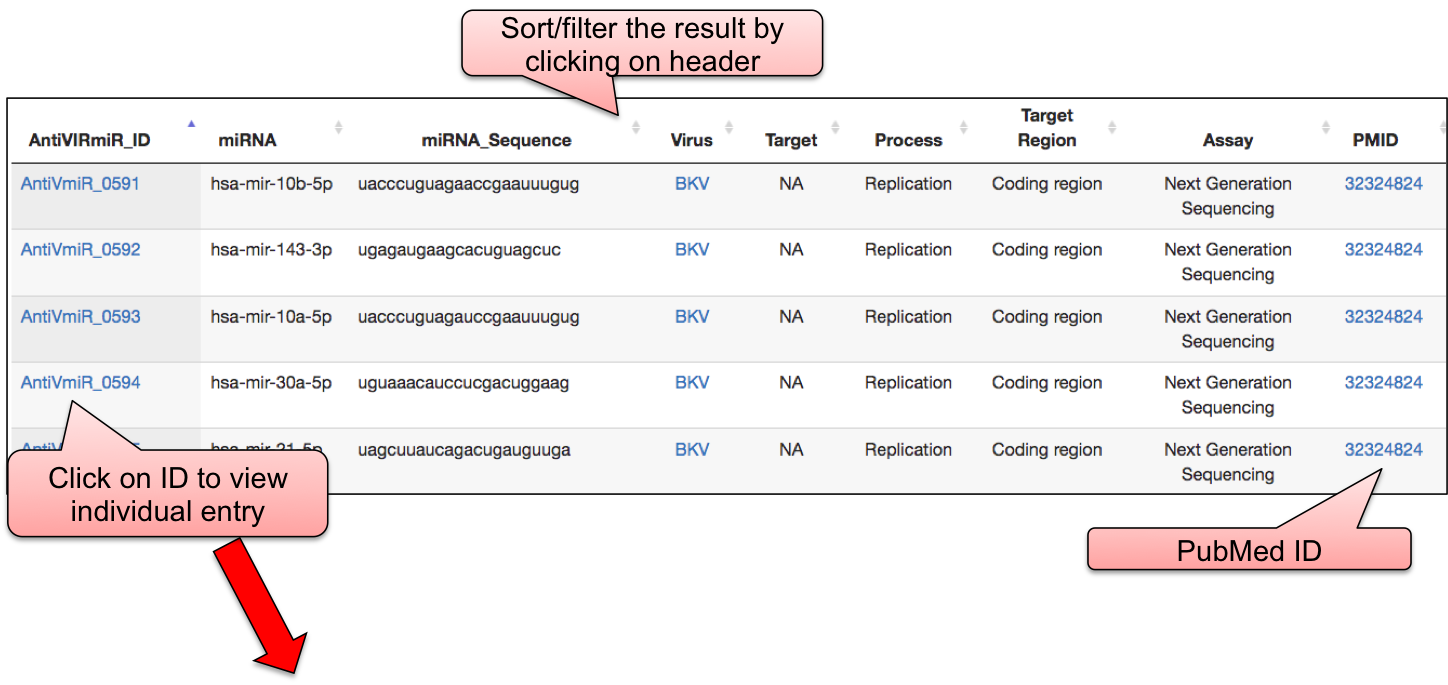
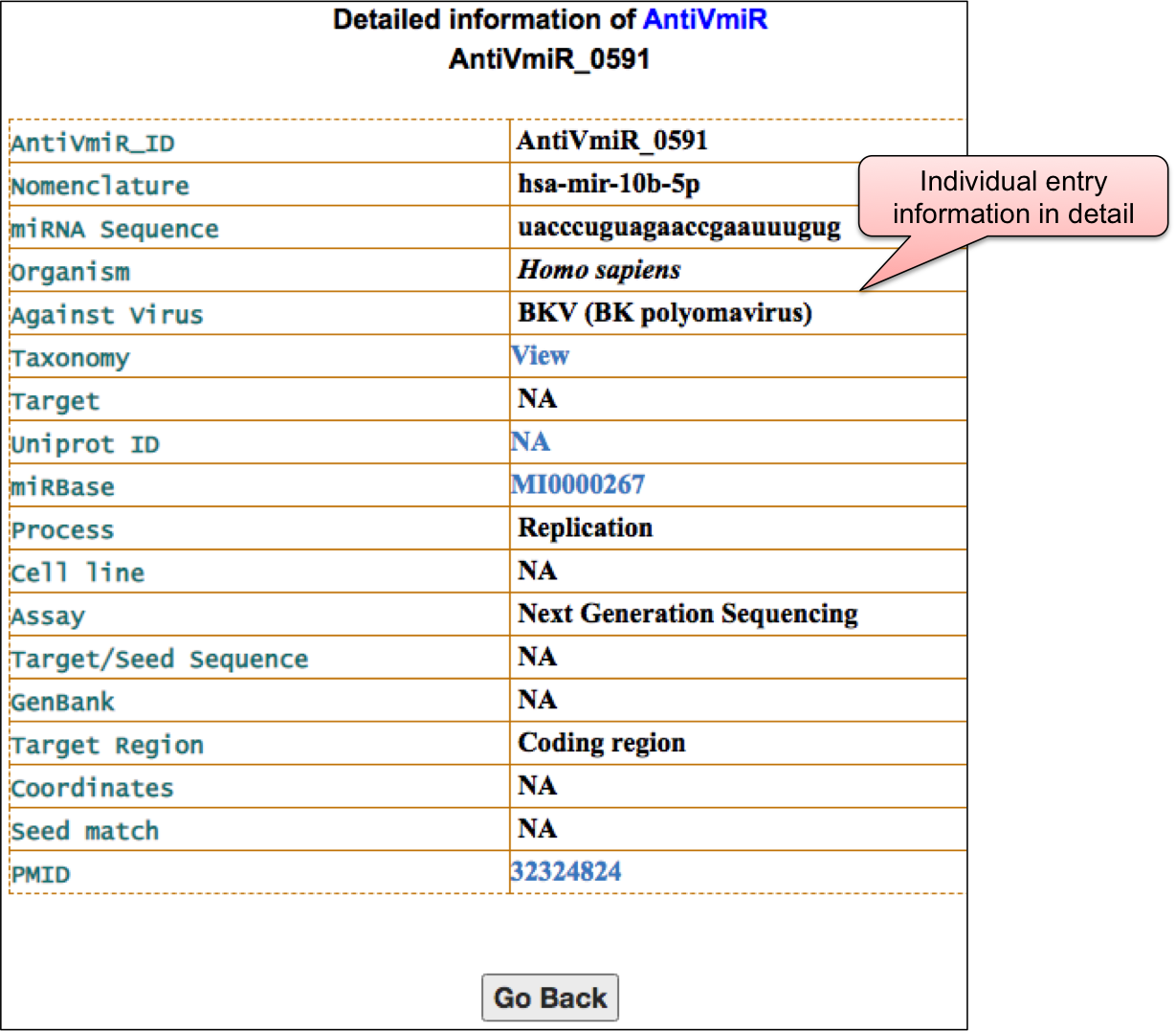
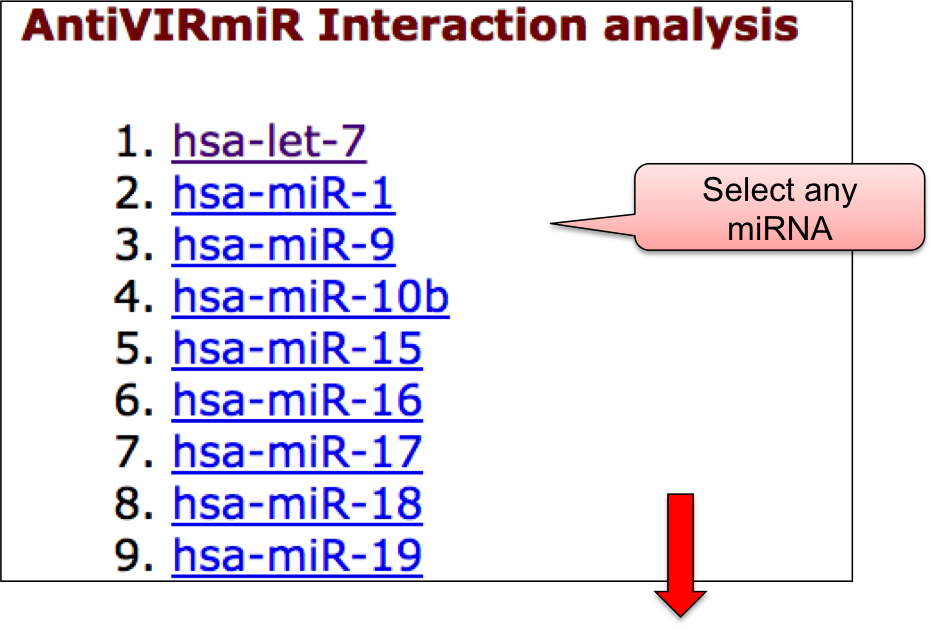
‘AntiVmiR’ webpage allowed user to have a glance at the list of viruses. The name of viruses are enlisted with the respective number of entries in the bracket. ‘AntiVmiR’ section incorporates 1642 entries for host miRNAs that show antiviral as well as proviral activity for 36 viruses. By clicking on the name of particluar virus, user can check the details of host miRNA that are tested against different viral infection.
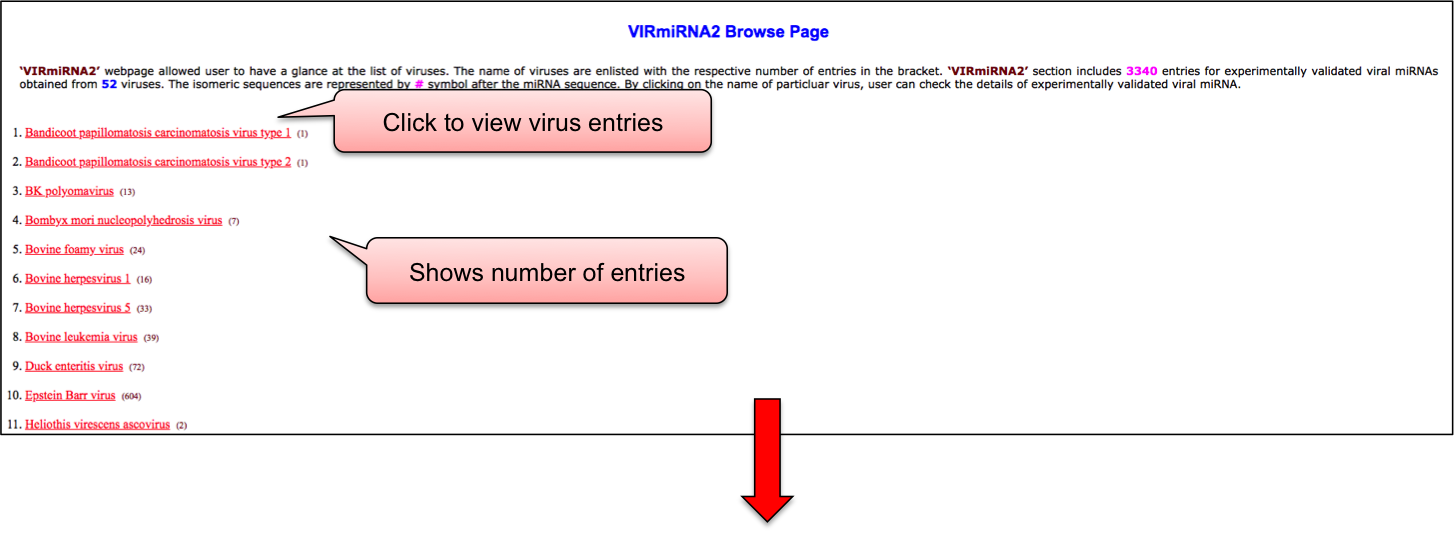
‘VIRmiRNA2’ browse page allowed user to have a glance at the list of viruses. The name of viruses are enlisted with the respective number of entries in the bracket. This section includes 3340 entries for experimentally validated viral miRNAs obtained from 56 viruses. By clicking on the name of particluar virus, user can check the details of experimentally validated viral miRNA.
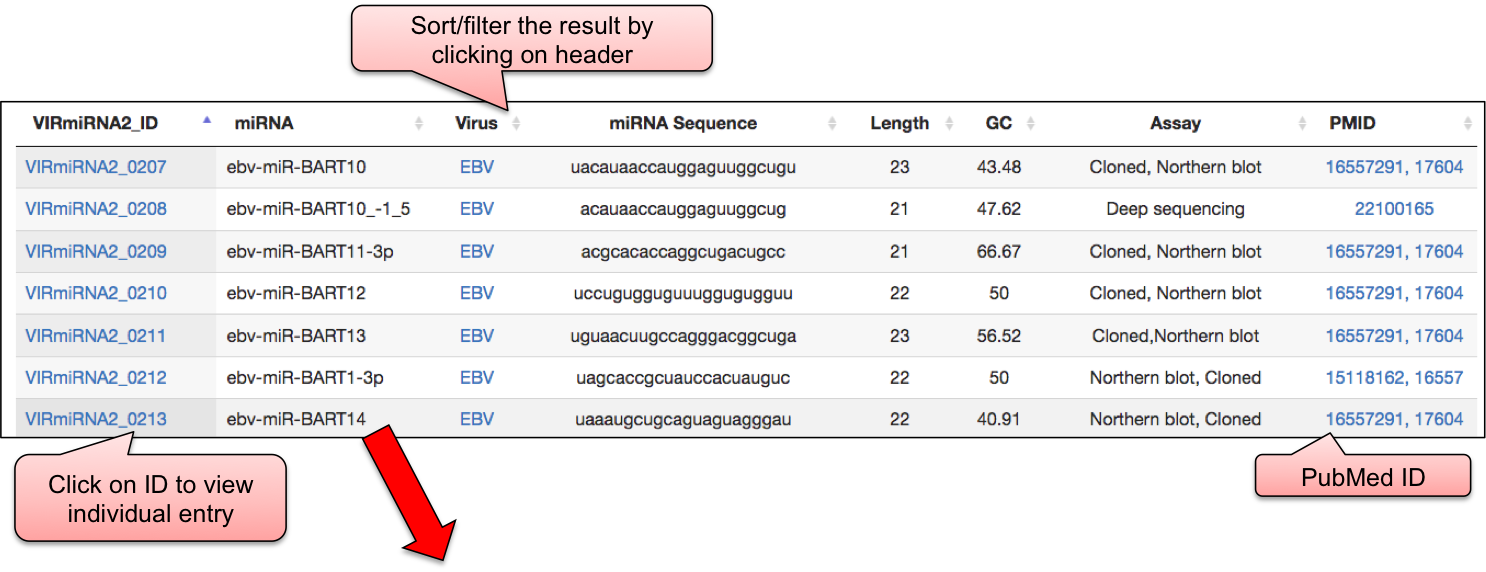
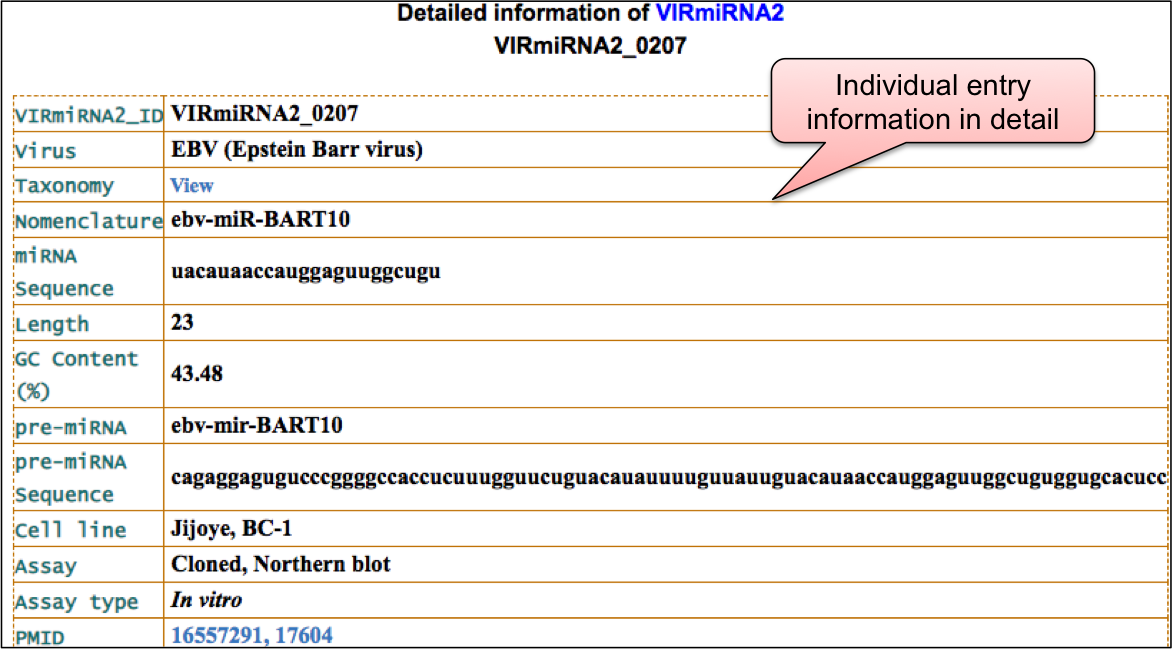
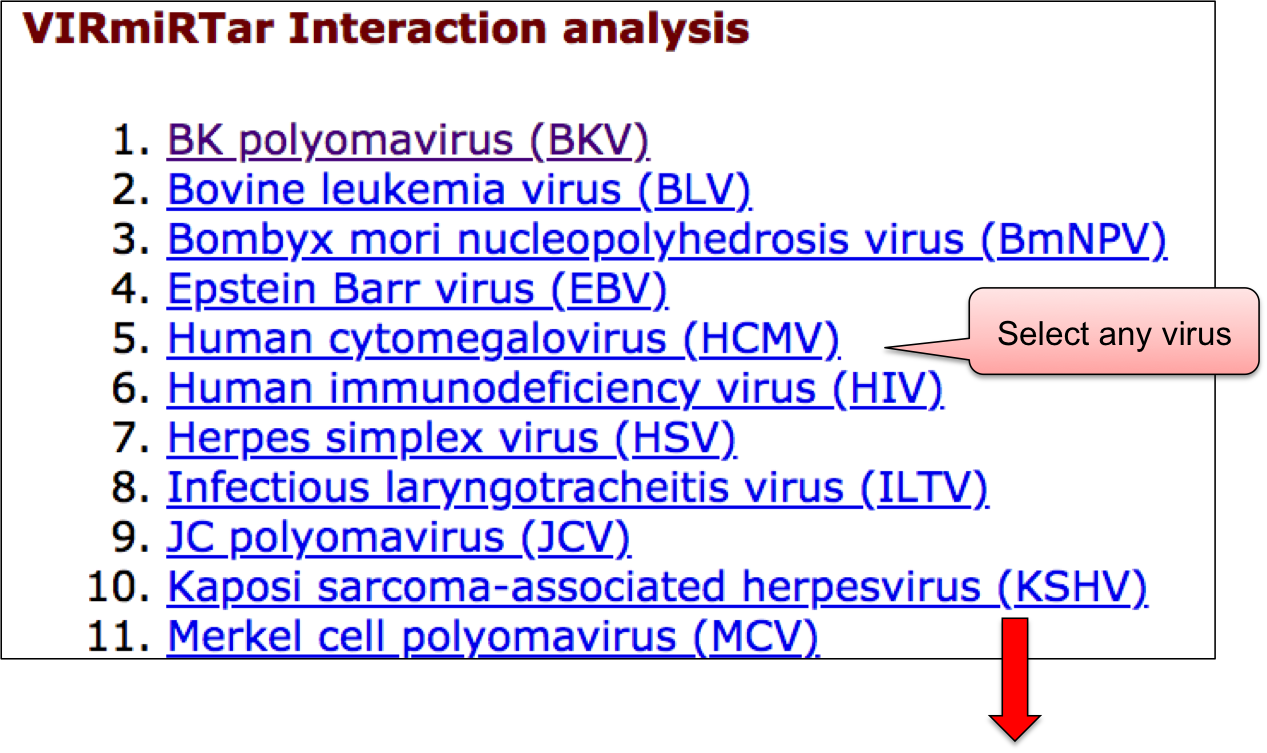
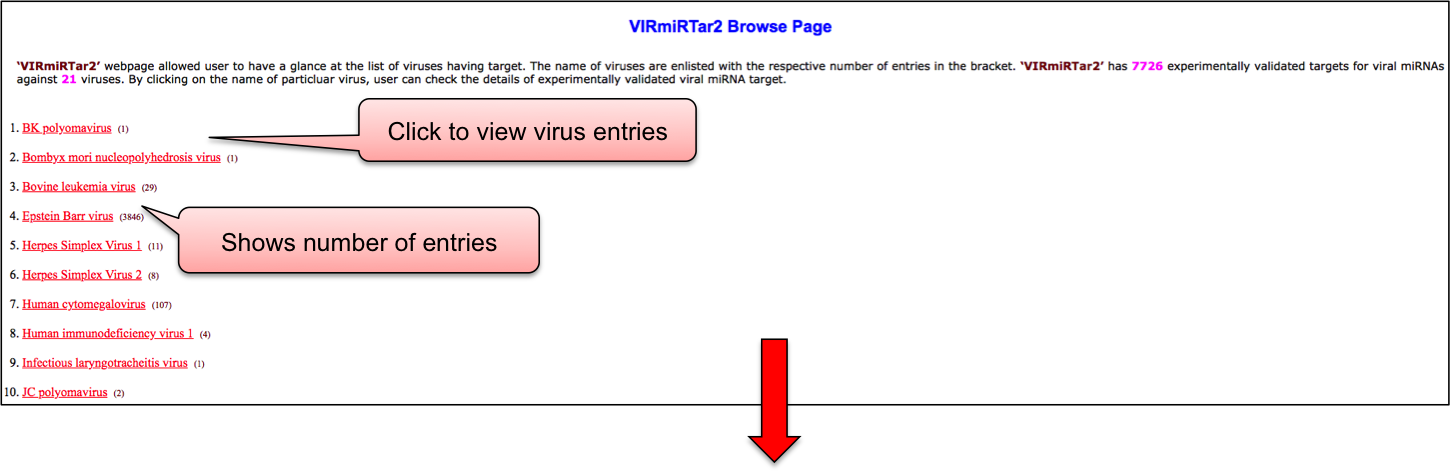
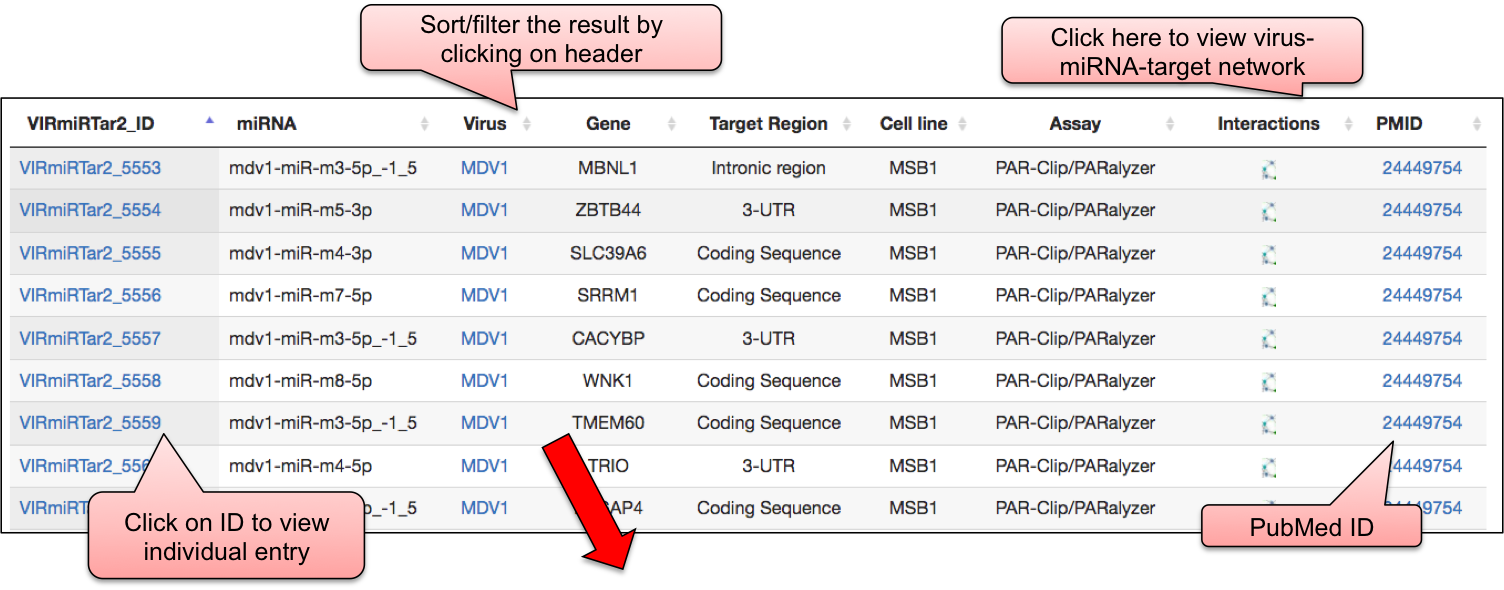
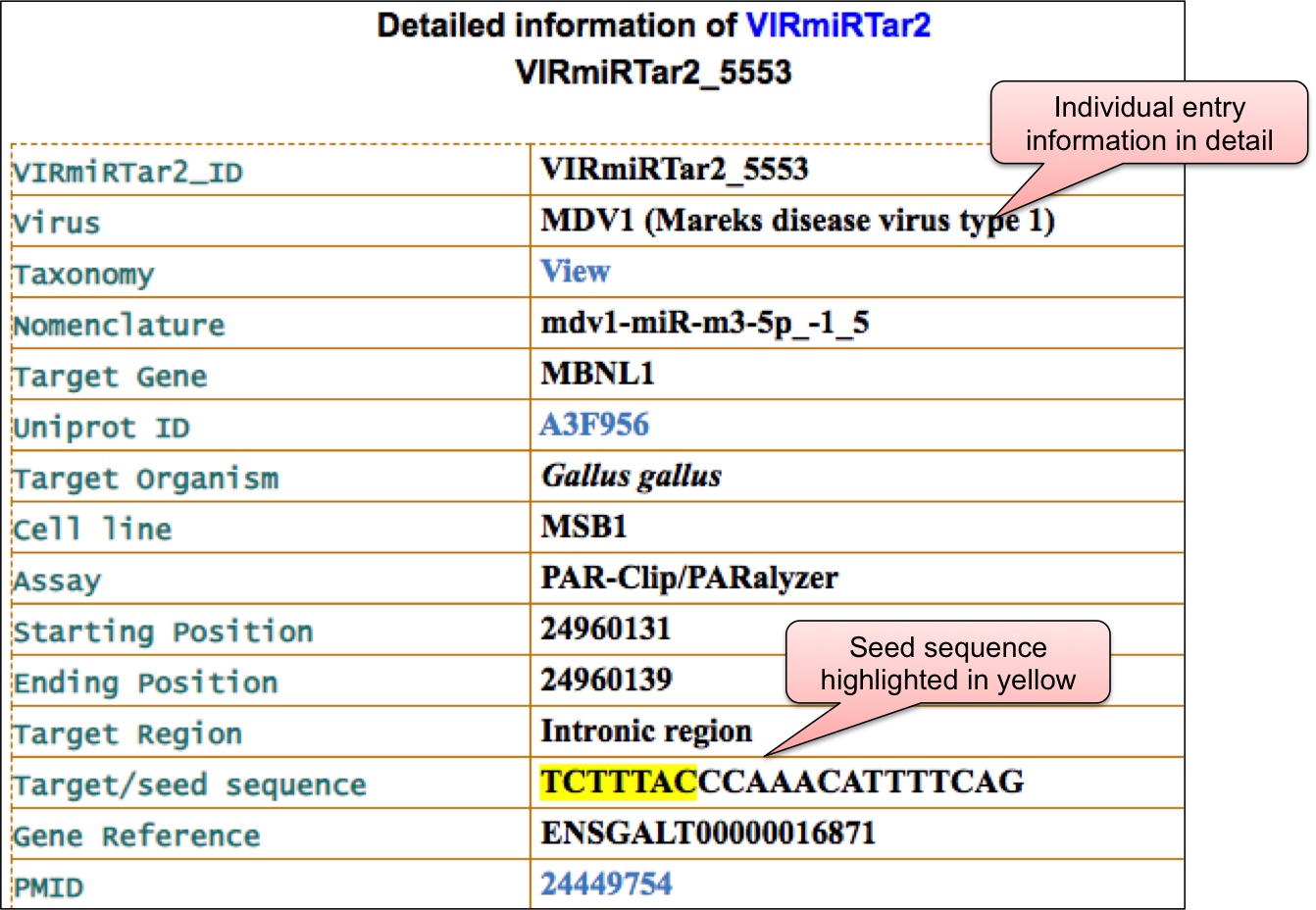
‘VIRmiRTar2’ webpage allowed user to have a glance at the list of viruses having target. The name of viruses are enlisted with the respective number of entries in the bracket. ‘VIRmiRTar2’ has 7726 experimentally validated targets for viral miRNAs against 21 viruses. By clicking on the name of particluar virus, user can check the details of experimentally validated viral miRNA target.
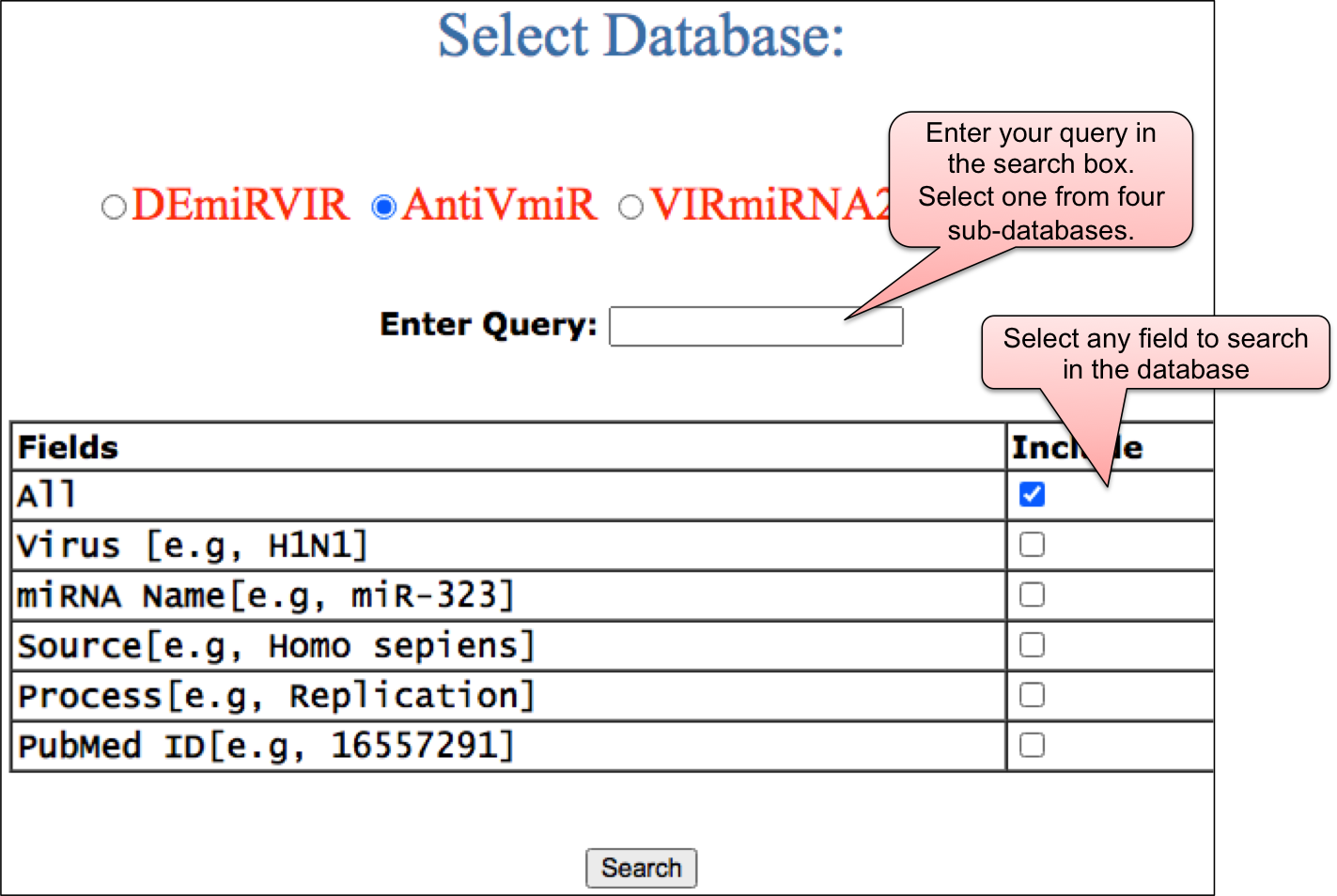
The search option is incorporated into four sub-databases - DEmiRVIR, AntiVmiR, VIRmiRNA2 and VIRmiRTar2. By selecting one option from four sub-databases, the user can enter the query in the box. This query search can be done in any of the given fields like virus name, miRNA name or miRNA sequence etc. or else user can select the “ALL” means query will be searched against all the fields.
AntiVIRmiR allow users to take advantage of different tools given below:
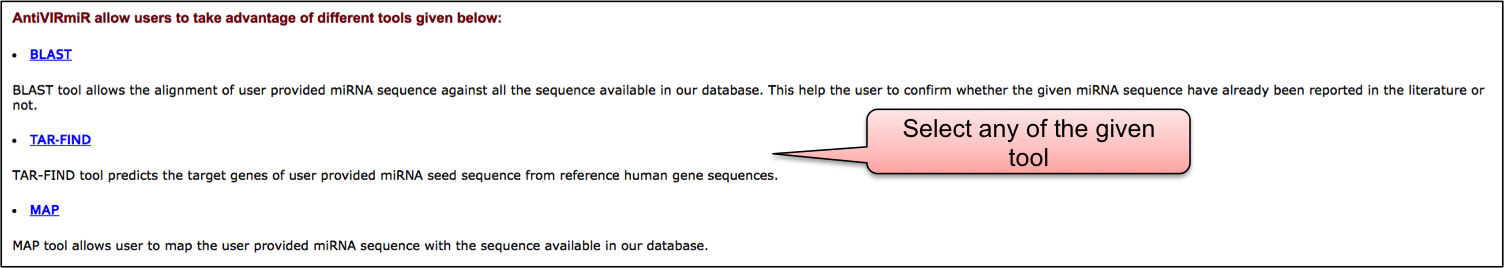
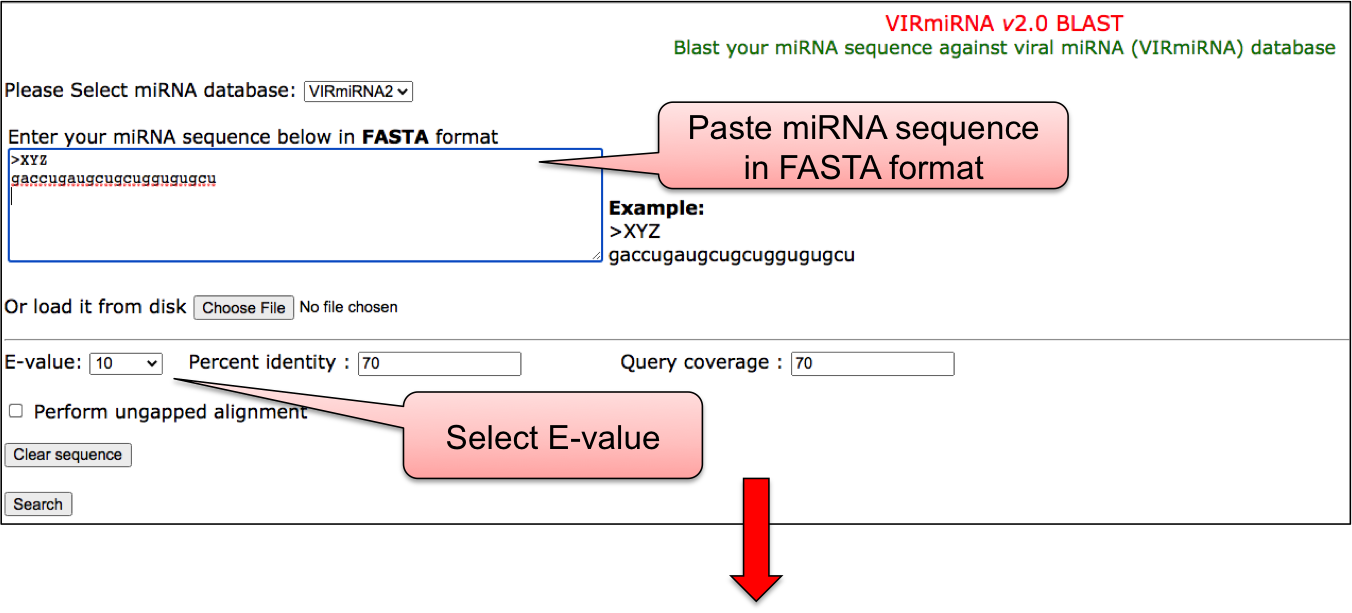
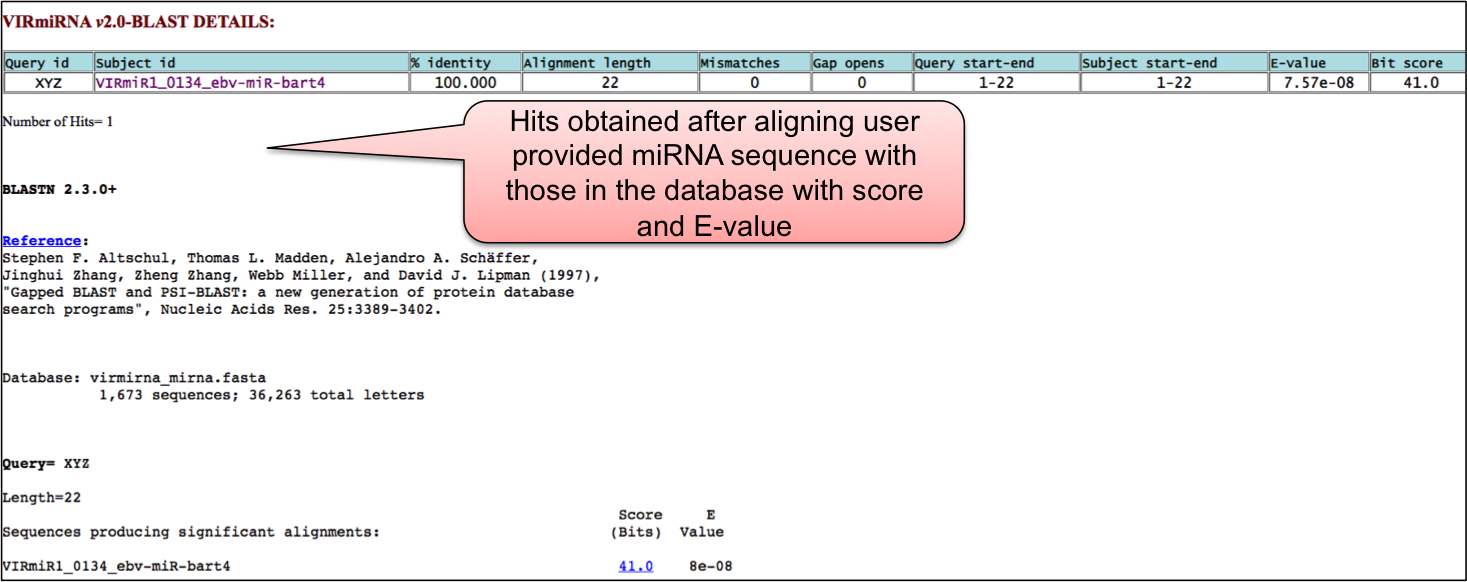
BLAST allows the alignment of user provided miRNA sequence against all the sequence available in our database. This help the user to confirm whether the given miRNA sequence have already been reported in the literature or not.
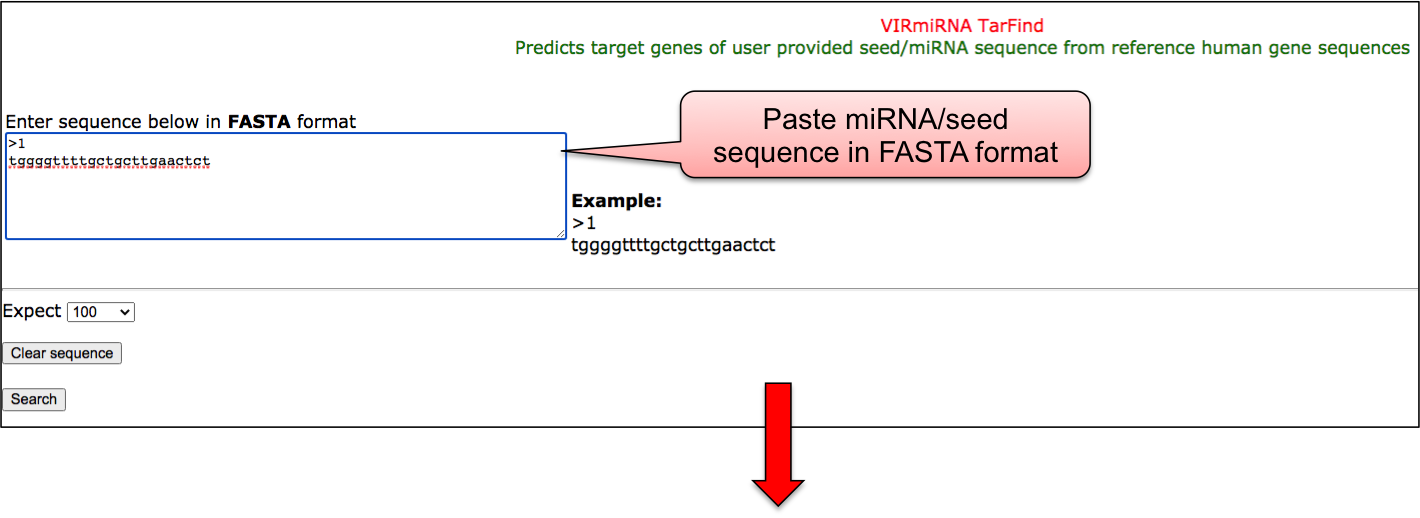
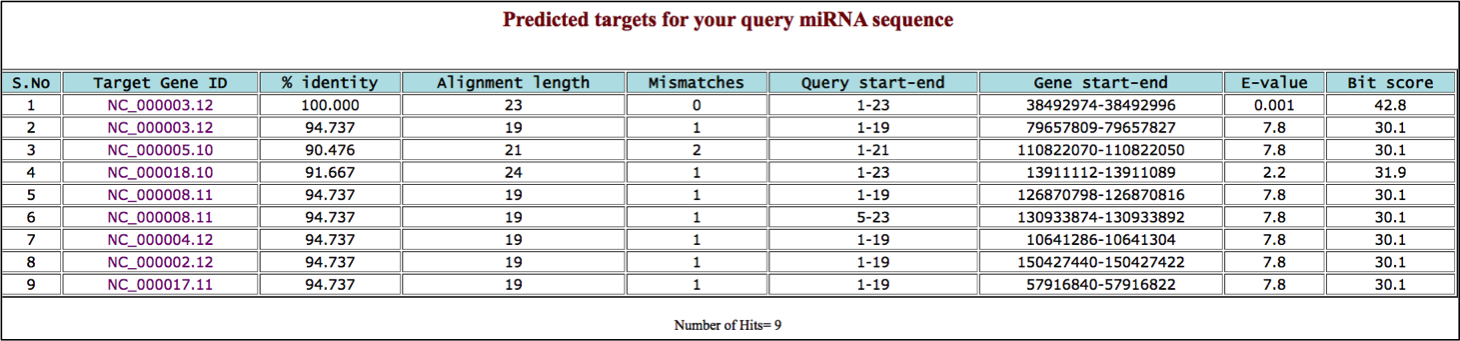
TAR-FIND tool predicts target genes of user provided miRNA seed sequence from reference human gene sequences.
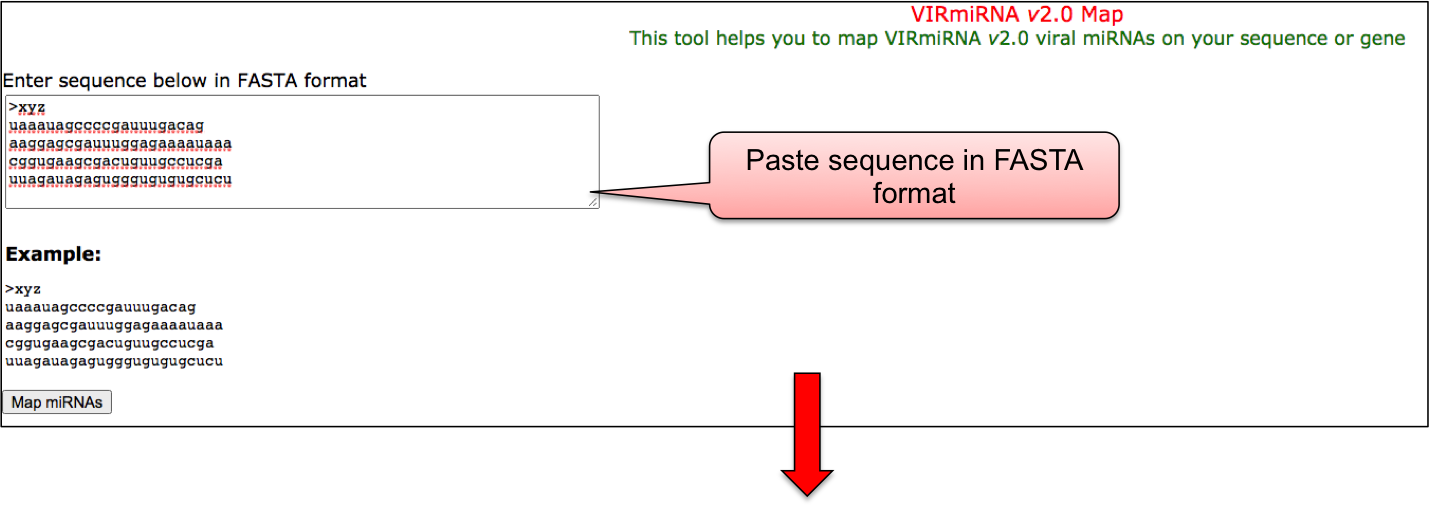
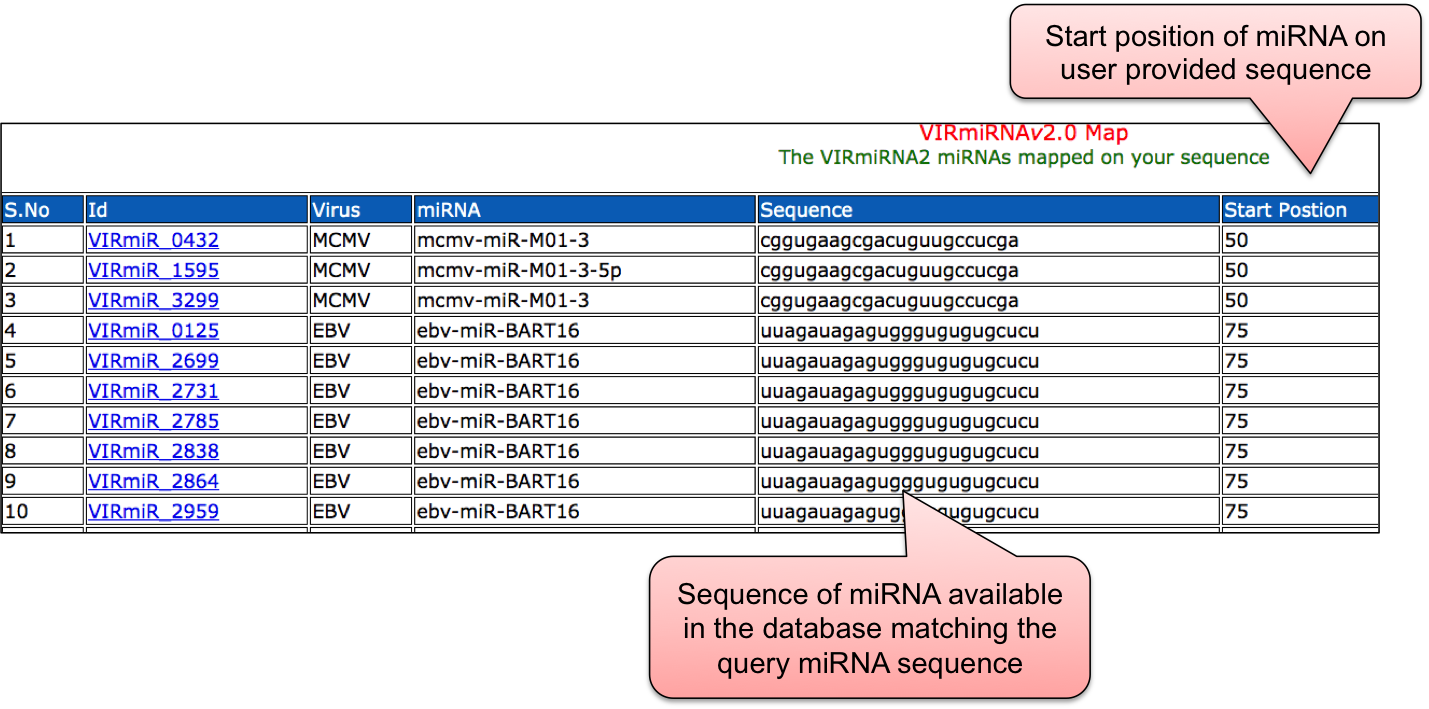
In Map tool, user can map given miRNA with those in our VIRmiRNA sub-database.
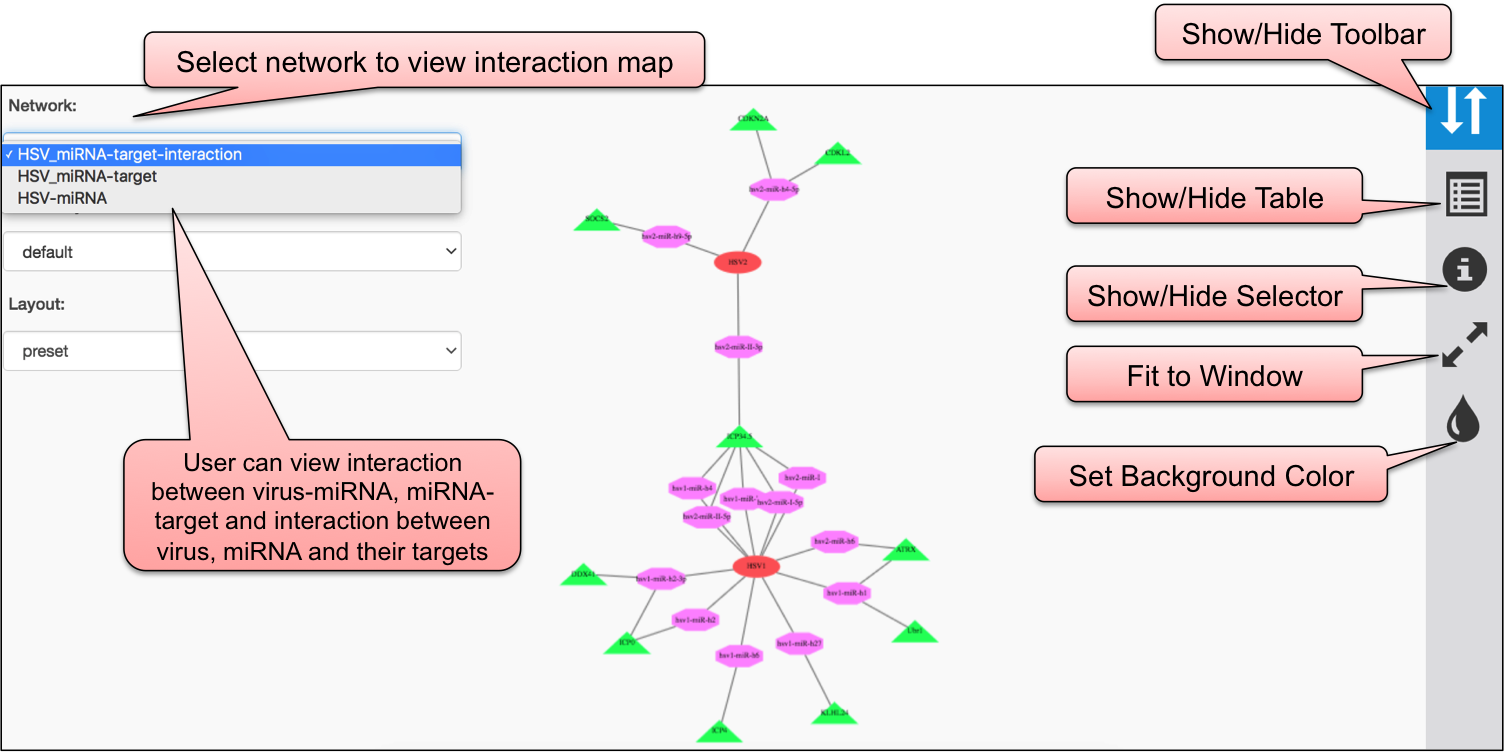
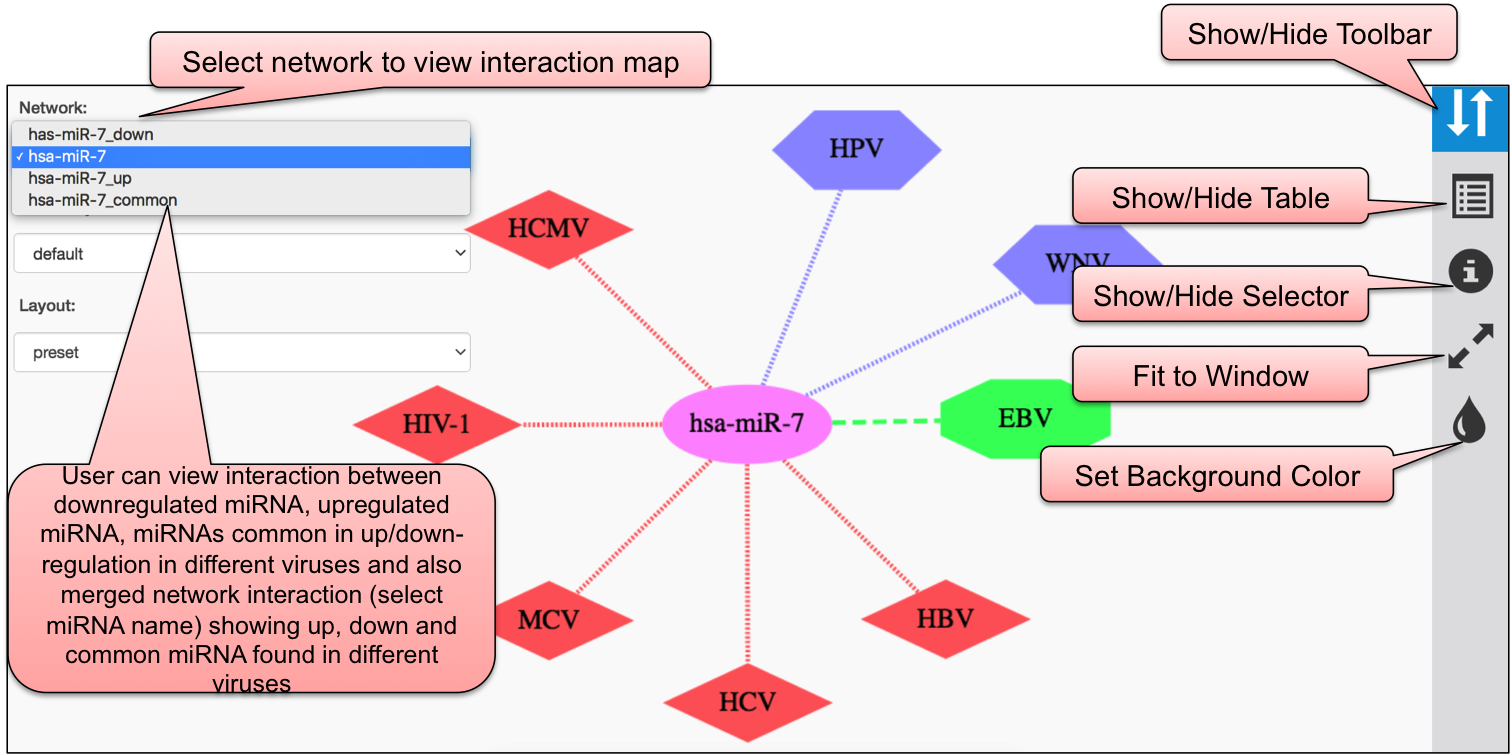
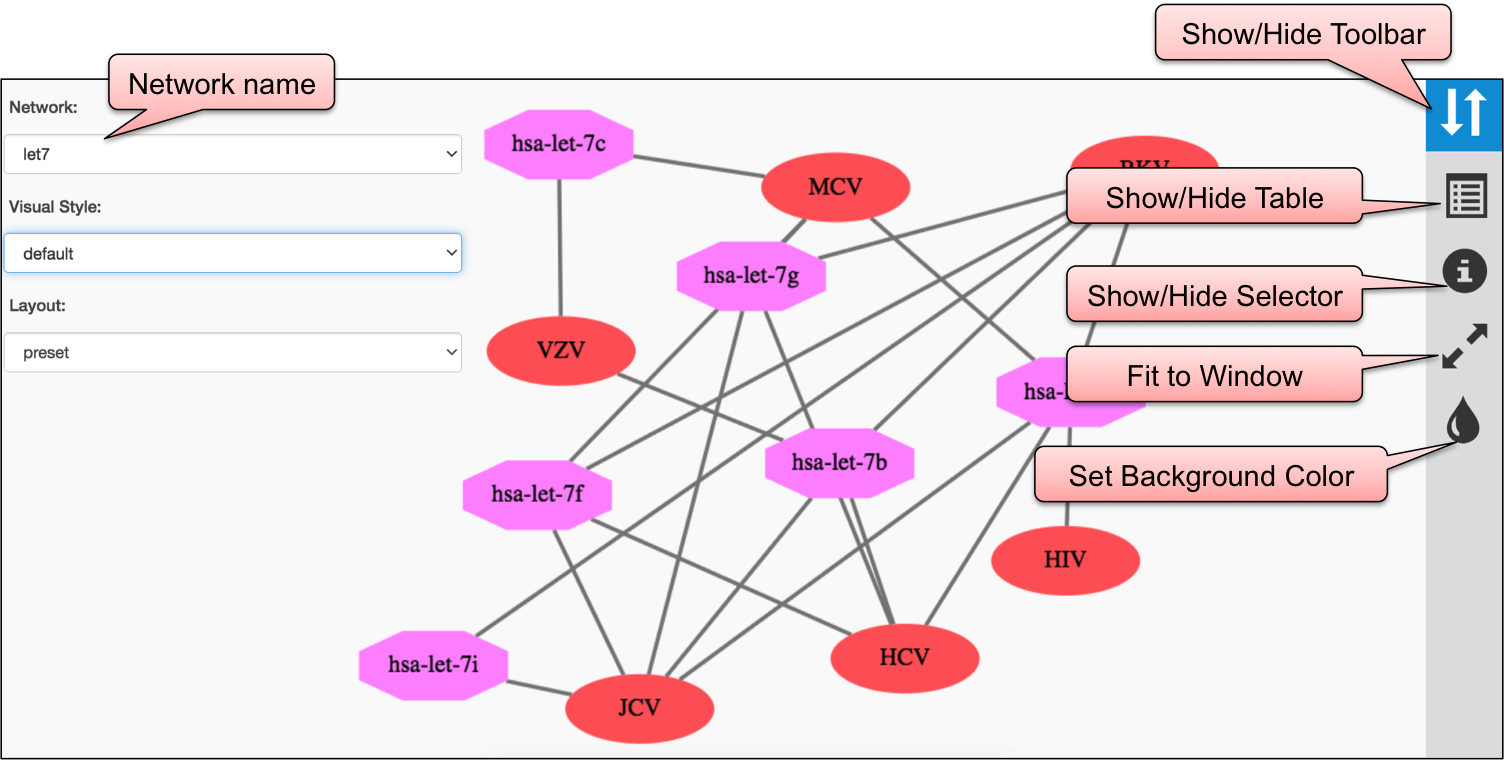
Using CYTOSCAPE software, we have developed interaction map of three sections, viz. DEmiRVIR, AntiVmiR and VIRmiRTar2. In DEmiRVIR, network is developed for host (human) miRNA that are upregulated, downregulated or both in different viruses. Networks can be explored by clicking on the individual analysis.In AntiVIRmiR, we have developed the interaction map for host miRNA found common in different viruses. In the VIRmiRTar2 section, network is developed for viral miRNAs with their target found in different viruses.
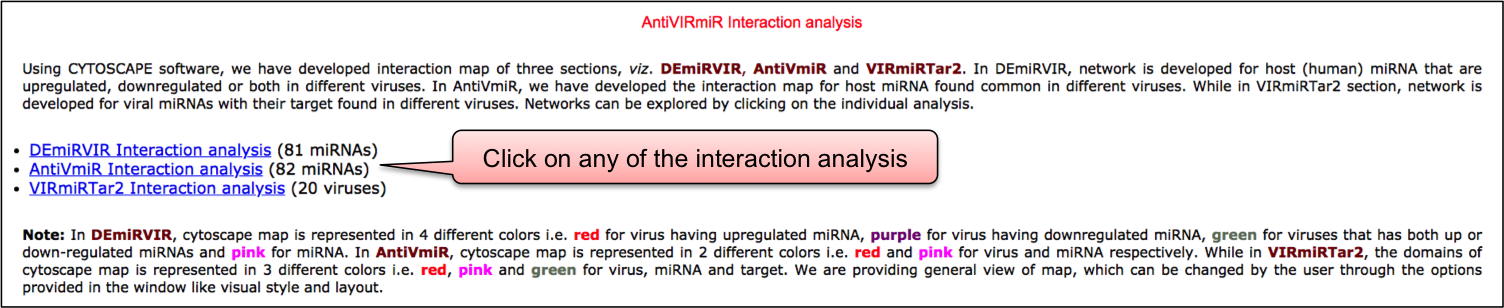
In DEmiRVIR, the network interaction is developed for 81 host/human miRNAs. Cytoscape map is represented in in 4 different colors i.e. red for virus having upregulated miRNA, purple for virus having downregulated miRNA, green for viruses that has both up or down-regulated miRNAs and pink for miRNA.
In AntiVmiR, the network interaction is developed for 82 host/human miRNAs. Cytoscape map is represented in 2 different colors i.e. red and pink for virus and miRNA respectively.
In VIRmiRTar2, the network interaction is developed for 20 viruses. The domains of cytoscape map is represented in 3 different colors i.e. red, pink and green for virus, miRNA and target.