Help
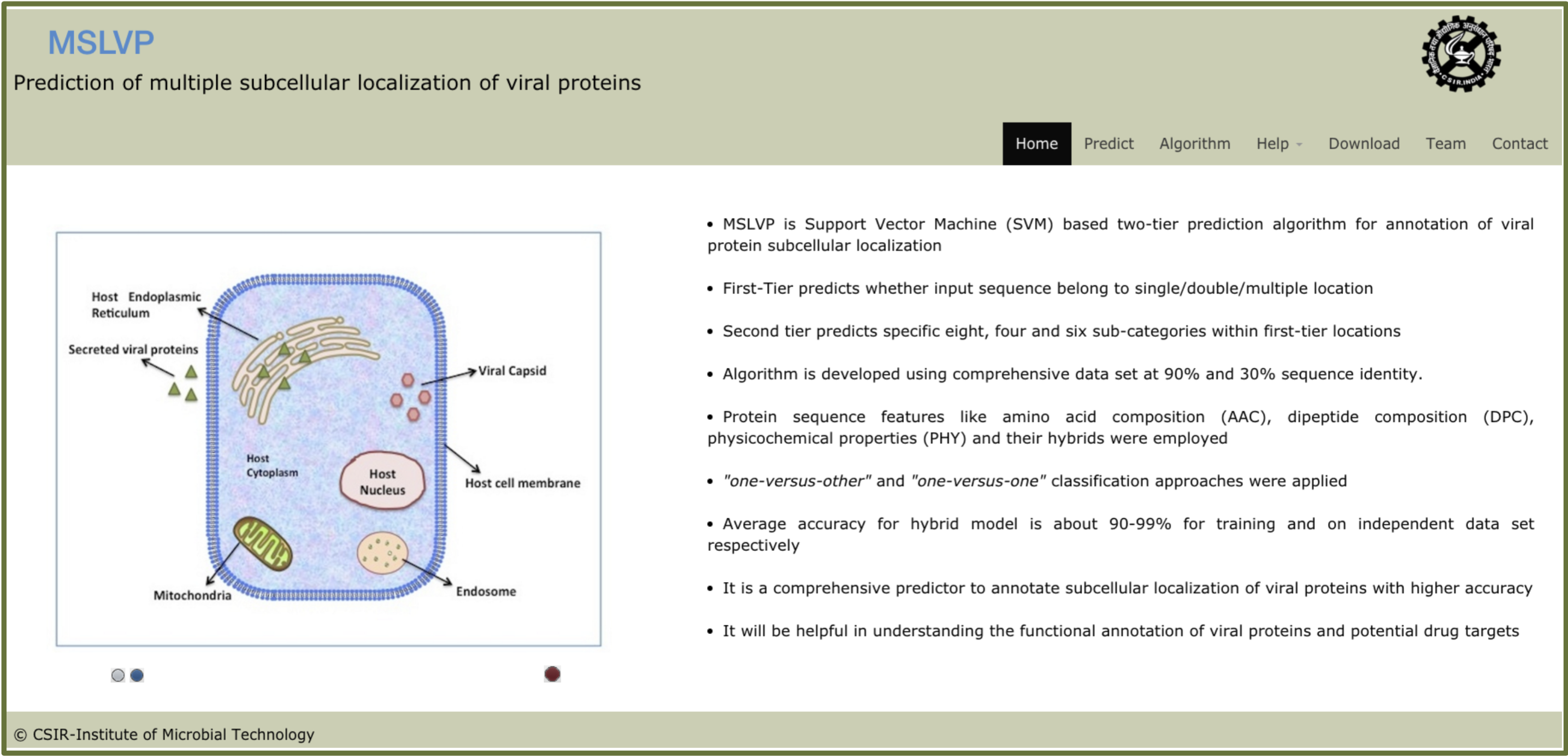
MSLVP is a two-tier based prediction algorithm for subcellular localization of viral proteins.A comprehensive data set were collected from UniProt and divided into three types of locations i.e. Single, Double and Multiple locations.
Result Interpretation
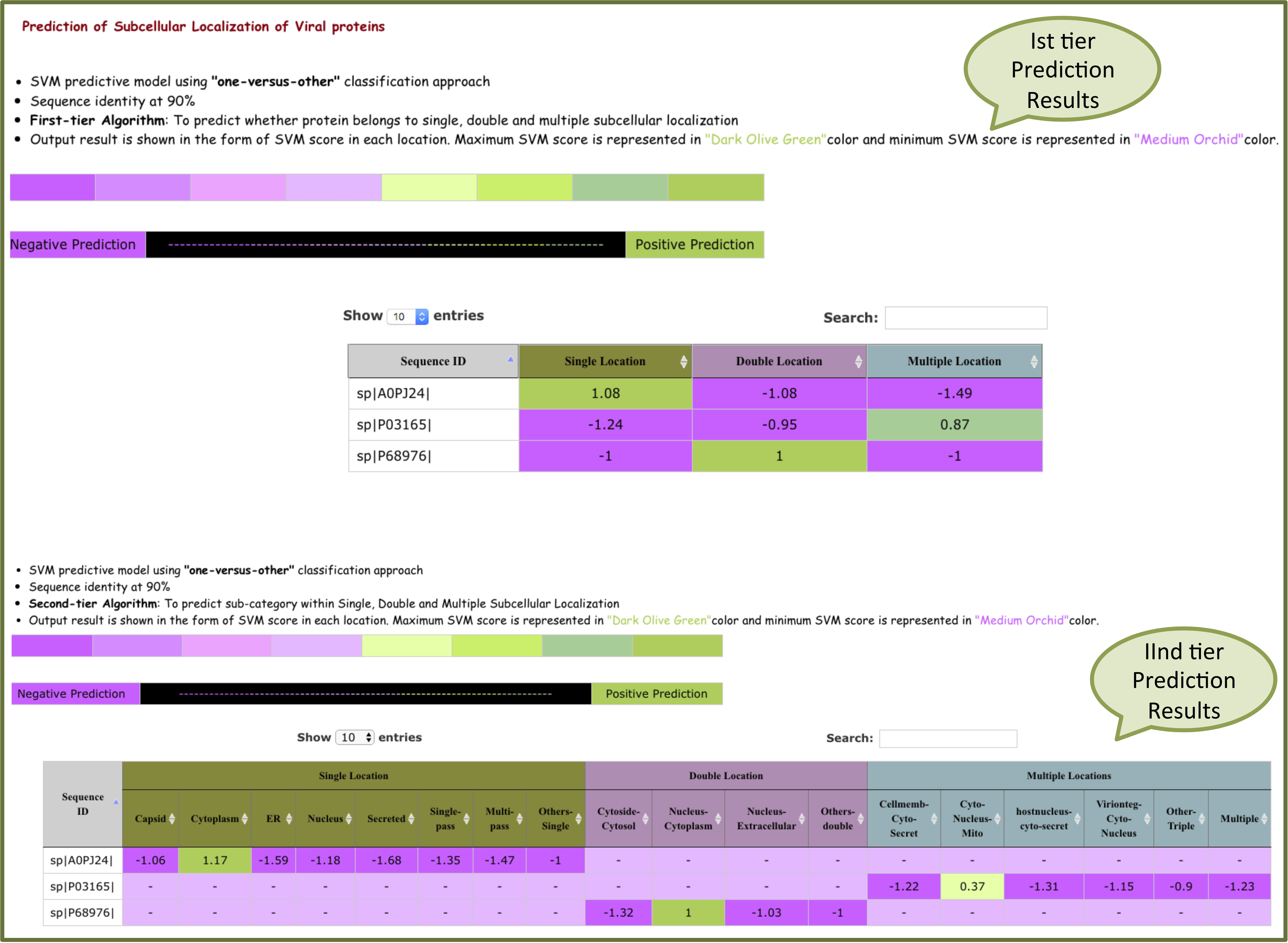
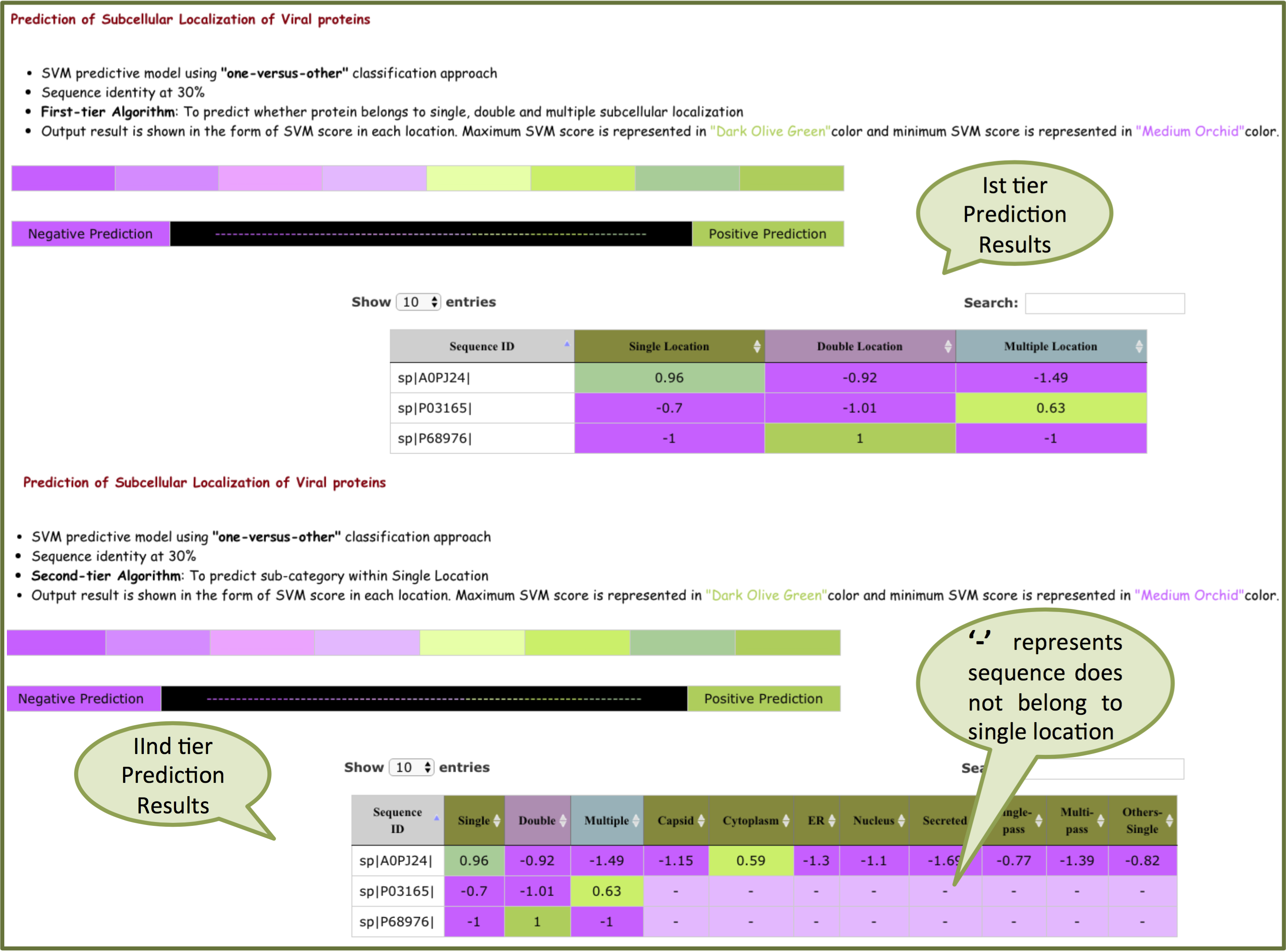
The results are analysed on the basis of color code shown below. color scheme is shown from negative to positive prediction based on SVM score. Higher the SVM score more the prediction confidence.
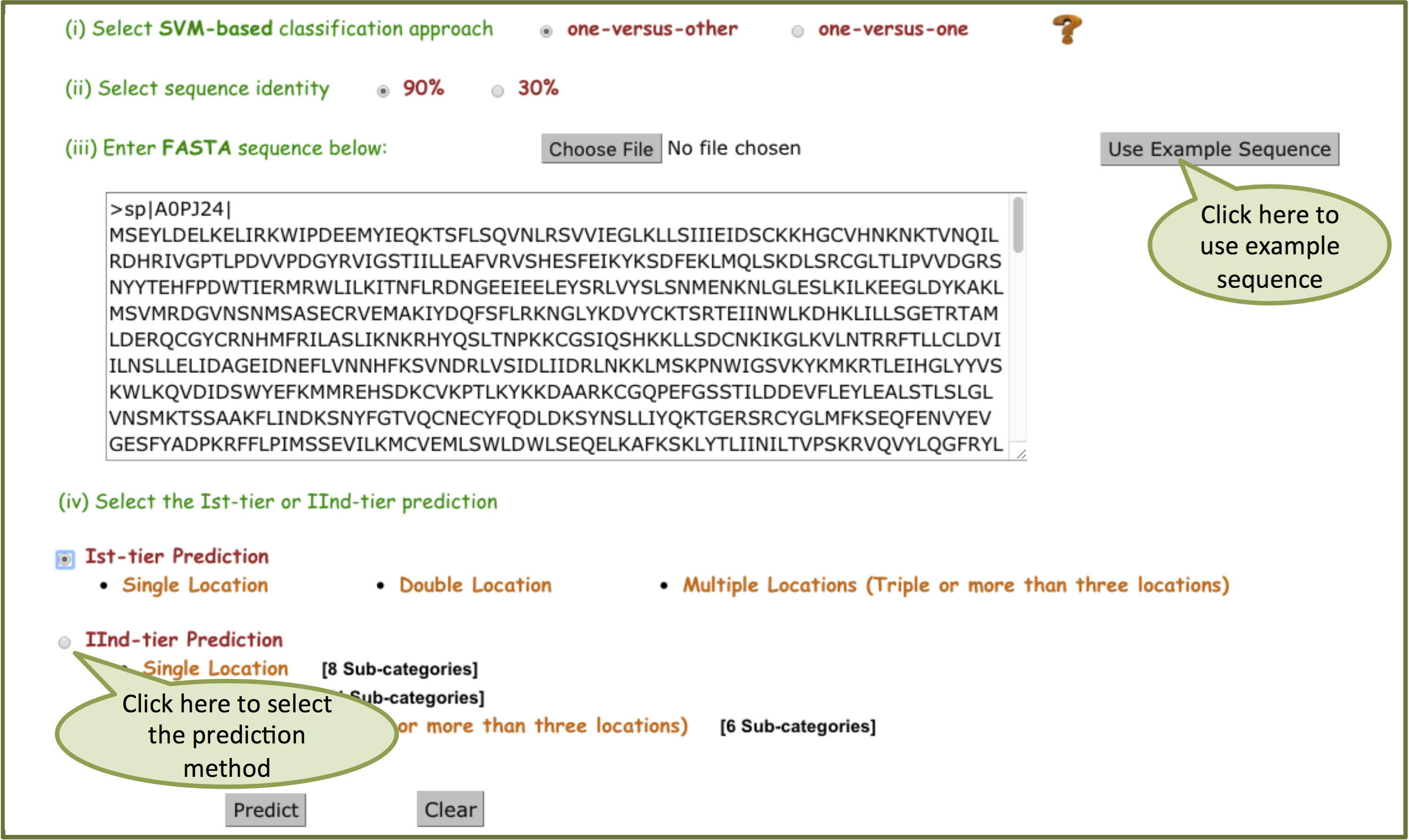
Paste your single or multiple sequence(s) in FASTA format in the text-box. User can remove the entries in the text-box by using the clear button. An "Example link" has been provided to help the users. By clicking on Example link, fasta sequences will be automatically filled in the text-box. User can select "one-versus-other" or "one-versus-one" classification. Select sequence identity at 90% or 30%. User can select Ist-tier and/or IInd-tier Prediction and click "Predict" button to make SCL predictions.
"one-versus-other" output will be displayed in the form of SVM scores. Higher the SVM score more the prediction confidence.
For 90% sequence identity, Ist -tier prediction predict score for single, double and multiple locations. Second-tier predicts the sub-category within single, double and multiple locations on the basis of score.
For 30% sequence identity, Ist -tier prediction predict score for single, double and multiple locations. If the sequence belongs to single location, second-tier predicts the sub-category within single locations on the basis of score. If sequence belongs to double o multiple location, "-" is displayed.
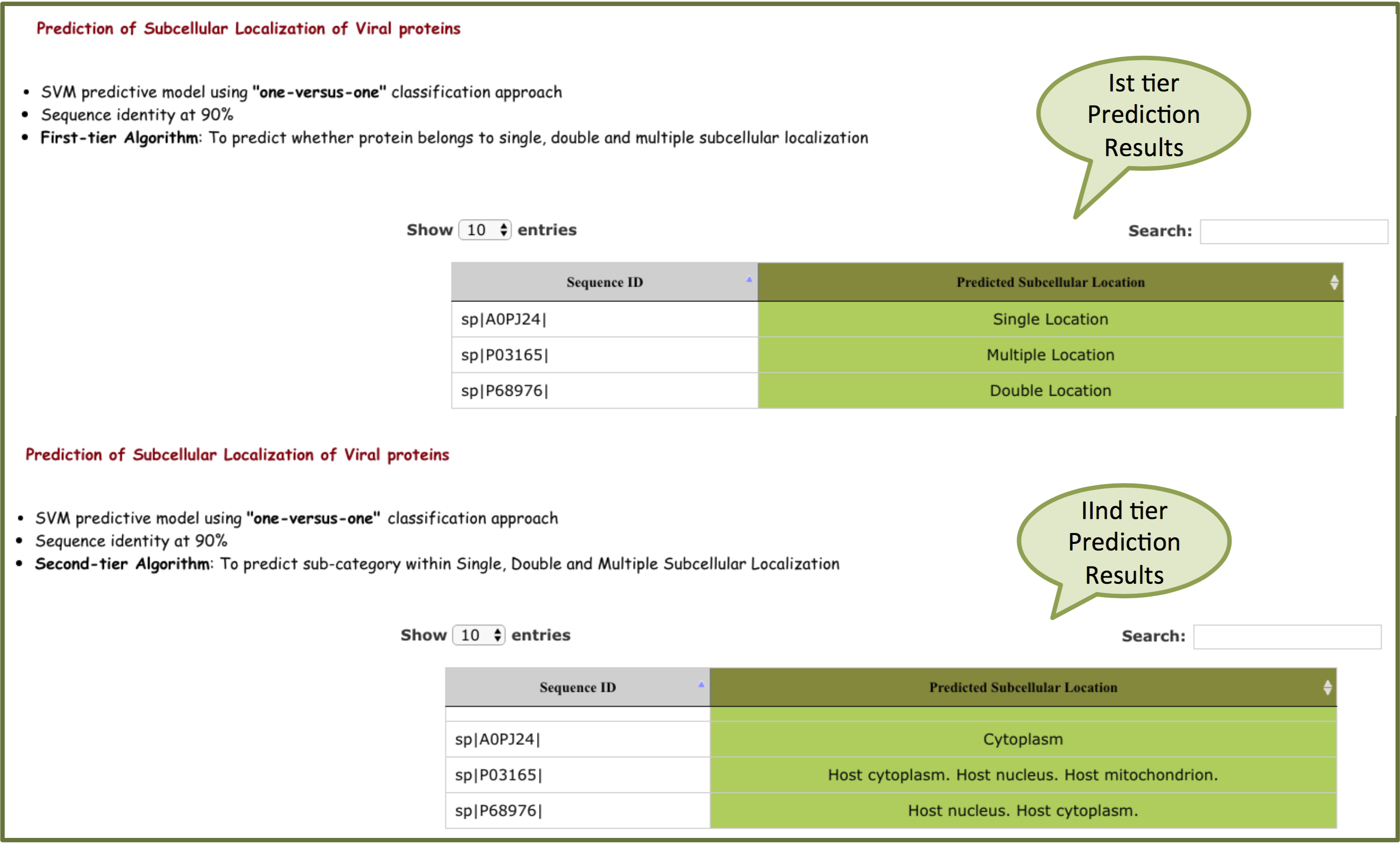
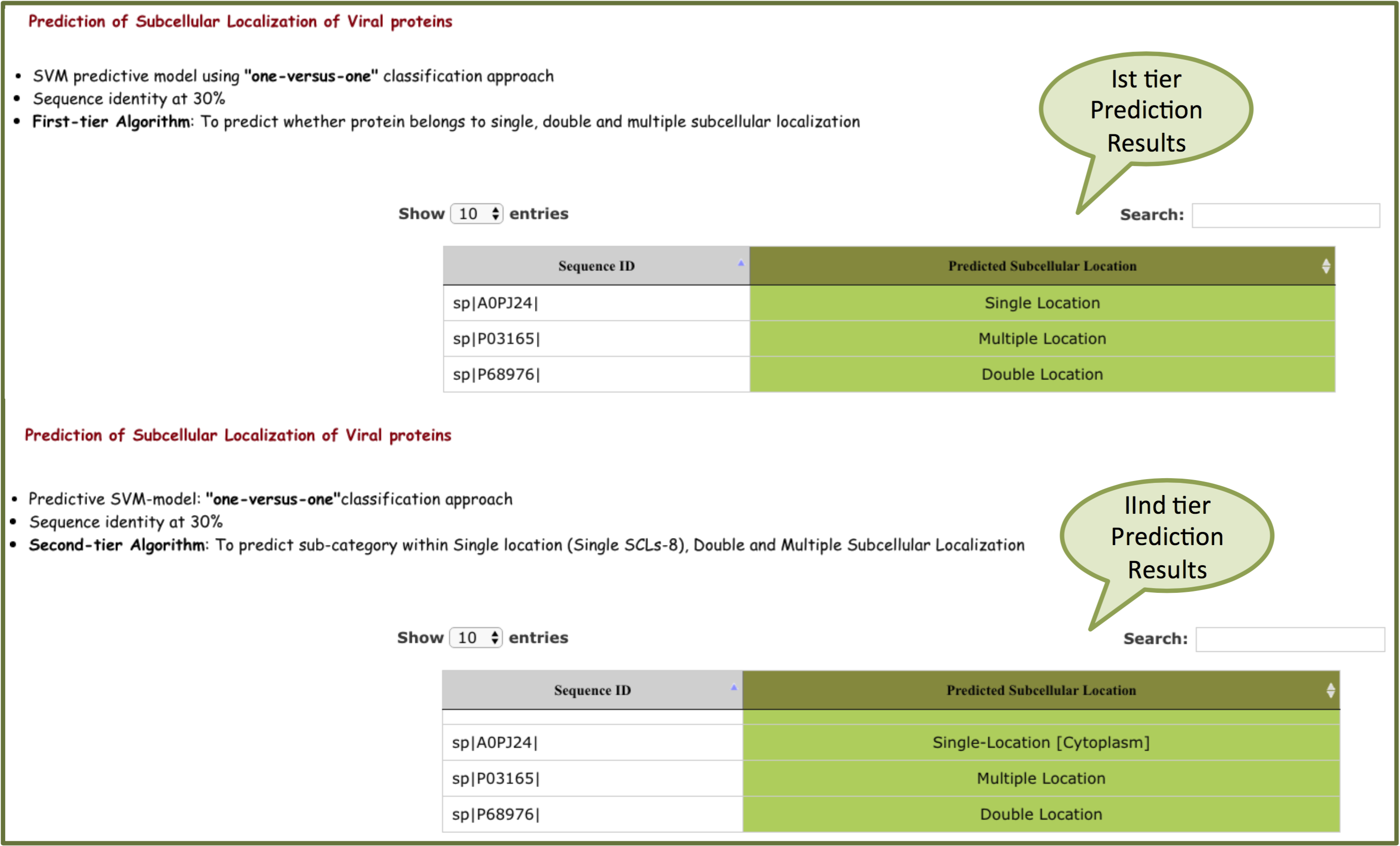
"one-versus-one" output will be displayed on the basis of predicted subcellular location.
For 90% sequence identity, Ist -tier prediction predict whether protein belongs to single, double and multiple locations. Second-tier predict whether protein belongs within the sub-category of single, double and multiple locations
For 30% sequence identity, Ist -tier prediction predict whether protein belongs to single, double and multiple locations. If the sequence belongs to single location, second-tier predicts the sub-category within single locations. If sequence belongs to double or multiple location, it will display only double or multiple location
__________
__________
__________
__________
__________
__________
__________
__________
Negative Prediction
--------------------------------------------------------------------------
Positive Prediction
MSLVP Input
MSLVP output
"one-versus-other"
"one-versus-one"