How to Use
anti-Ebola web server have been integrated using recursive regression based Quantitative structure–activity relationship (QSAR) models. It is freely available and easy to use.
-
1. SVM based anti-Ebola inhibitor
2. RF based anti-Ebola inhibitor
3. ANN based anti-Ebola inhibitor
4. Check the Job status
5. Format conversion tool
SVM based anti-Ebola inhibitor
anti-Ebola webportal contains Quantitative structure–activity relationship (QSAR) based prediction algorithm for predicting Ebola inhibition potential of chemical(s) using our Support Vector Machine (SVM) prediction model. The input will be provided in the form of .sdf format as single or multiple. The output would be displayed as tabulated format with Chemical ID, SMILES, Inhibition efficiency, Important descriptors, and structures of query chemical(s).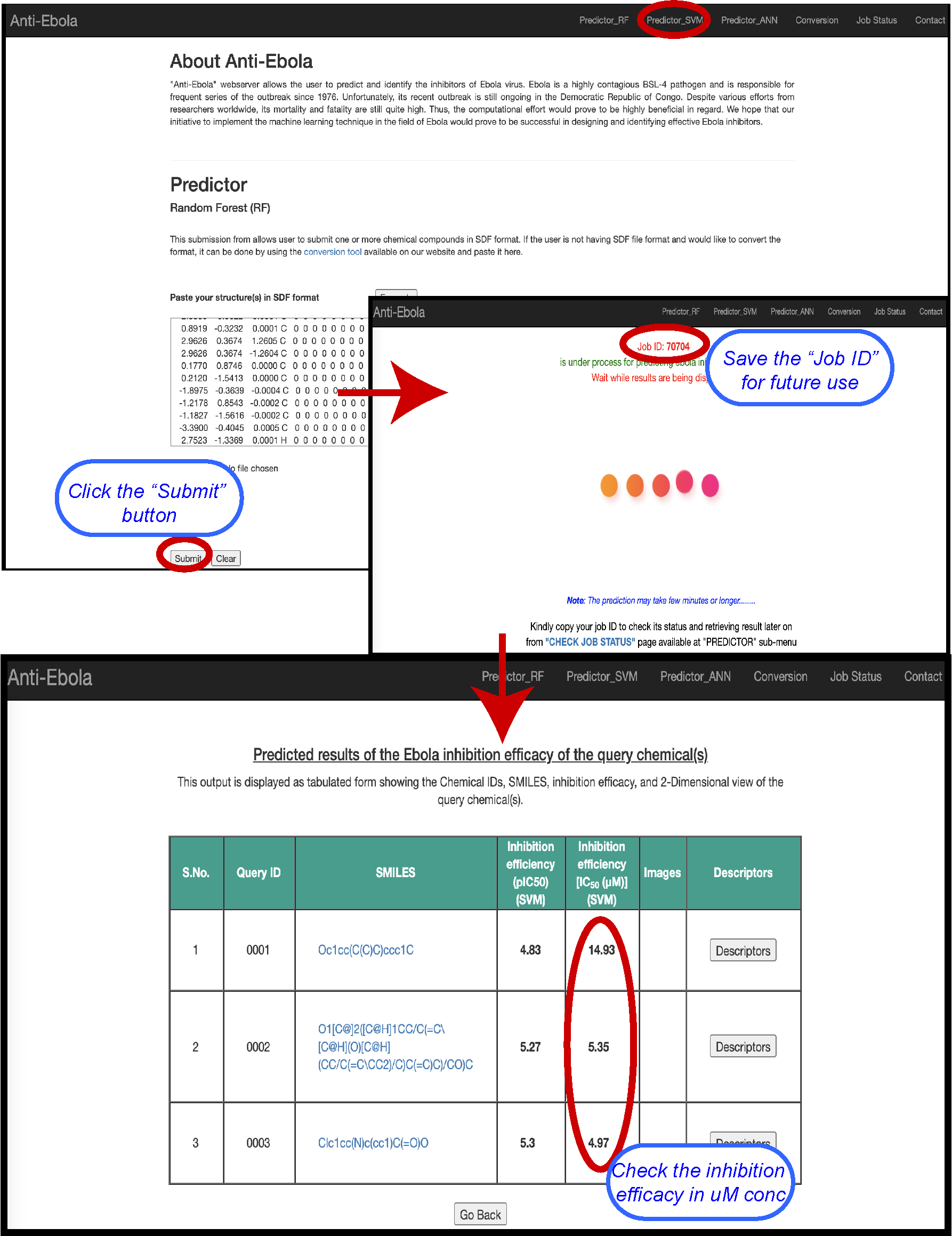
RF based anti-Ebola inhibitor
anti-Ebola webportal contains Quantitative structure–activity relationship (QSAR) based prediction algorithm for predicting Ebola inhibition potential of chemical(s) using our Random Forest (RF) prediction model. The input will be provided in the form of .sdf format as single or multiple. The output would be displayed as tabulated format with Chemical ID, SMILES, Inhibition efficiency, Important descriptors, and structures of query chemical(s).
ANN based anti-Ebola inhibitor
anti-Ebola webportal contains Quantitative structure–activity relationship (QSAR) based prediction algorithm for predicting Ebola inhibition potential of chemical(s) using our Artificial Neural Network (ANN) prediction model. The input will be provided in the form of .sdf format as single or multiple. The output would be displayed as tabulated format with Chemical ID, SMILES, Inhibition efficiency, Important descriptors, and structures of query chemical(s).
Check the Job status
As the completion of a job requires 2-5 min. So, user can save the ‘job id’ and retrieve it by using the Job Status web page.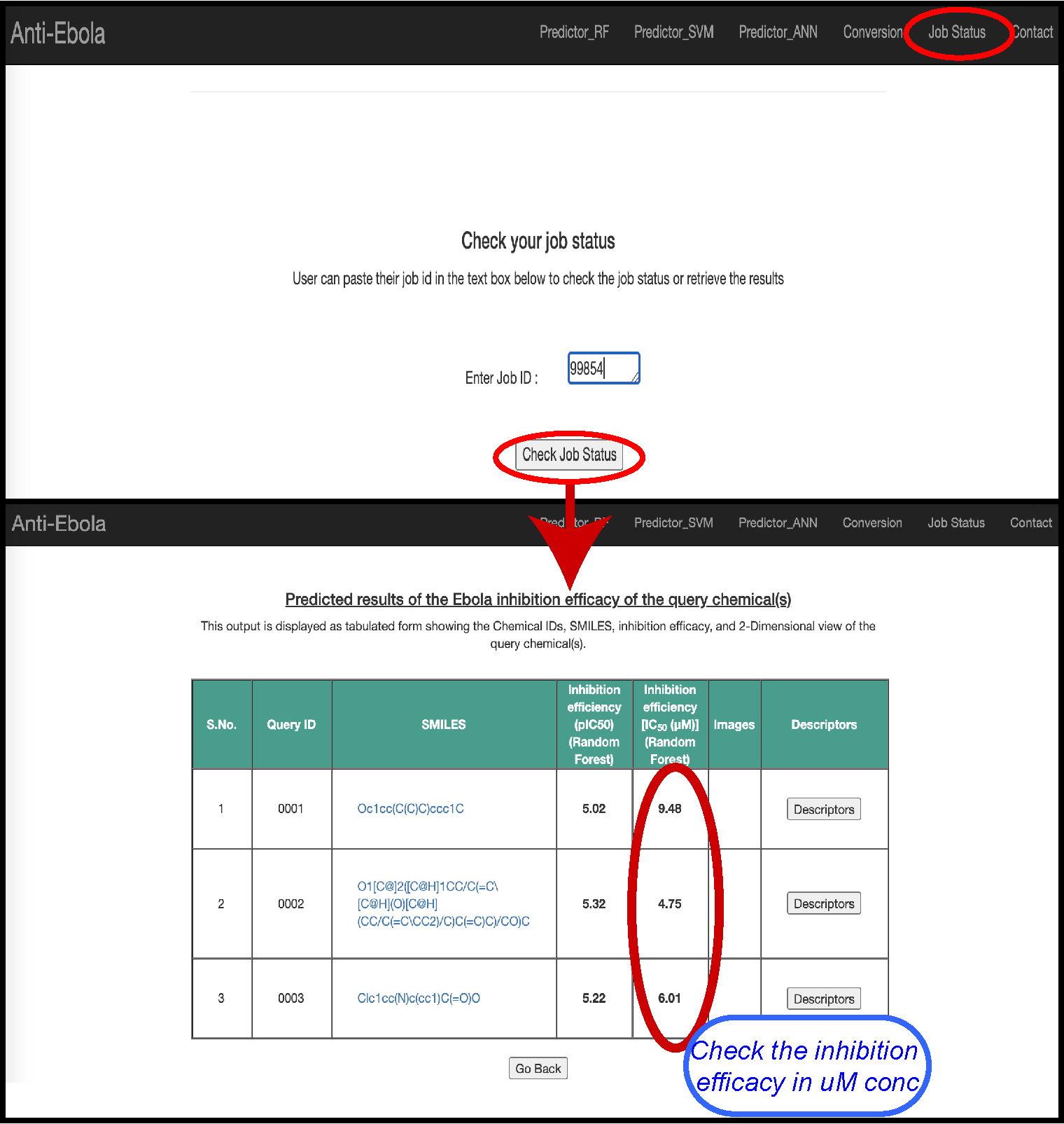
Format conversion tool
anti-Ebola webportal contains a Convertor tool with the ability to convert any chemical structure into SMILES, .SDF, .MOL format. The input will be provided in the form of chemical structure, where the user(s) can draw or paste the query structure in JSME editor. The output would be displayed as tabulated format with information of SMILES, .SDF, .MOL and chemical structure.