Help Page
This web page help the user to take a glance as how to use the "anti-Nipah" web server
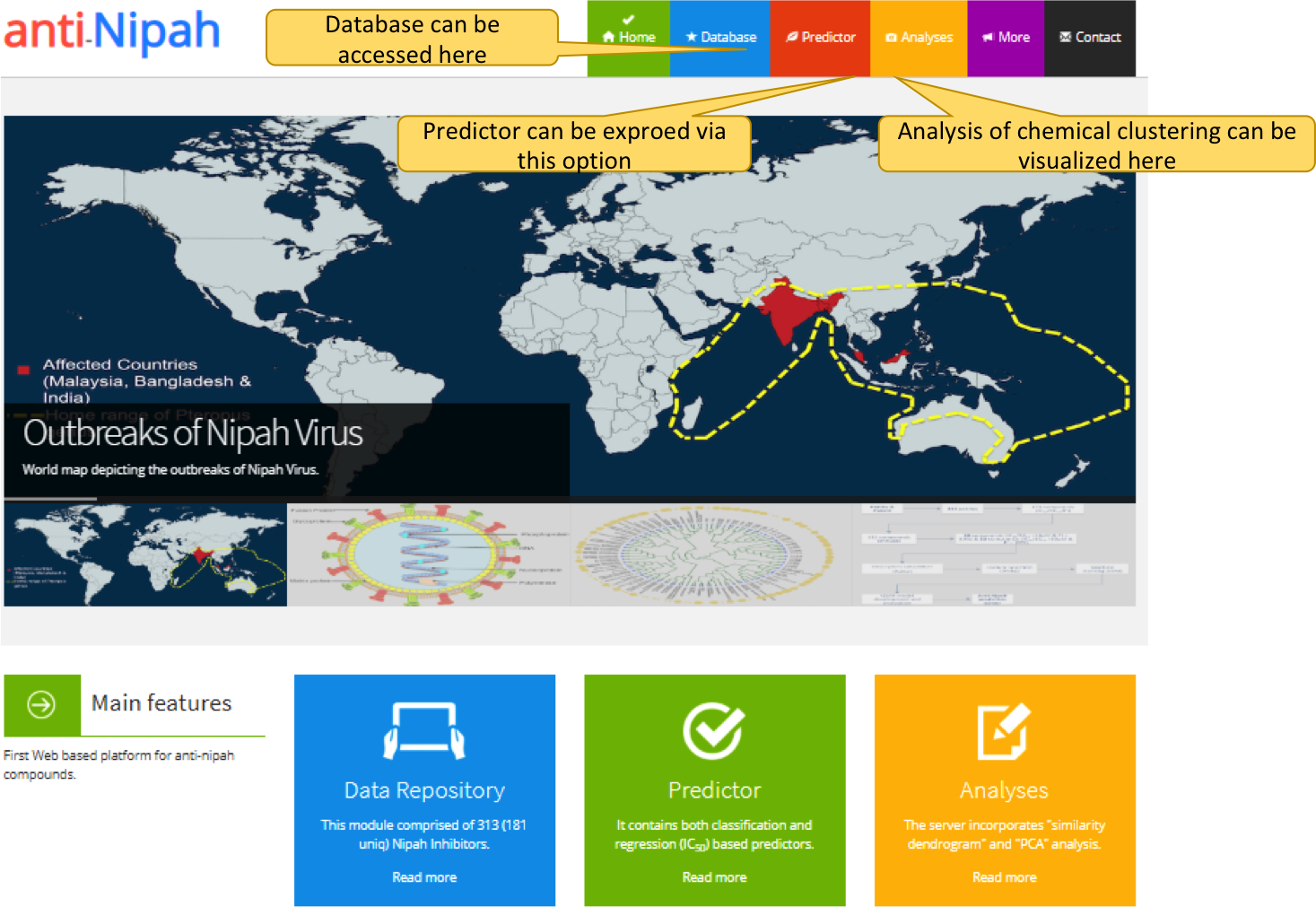
1. Anti-Nipah home page
"anti-Nipah" home page allows the user to explore the anti-Nipah compounds via three modules i.e. database, predictor and analyses
2. Browse page
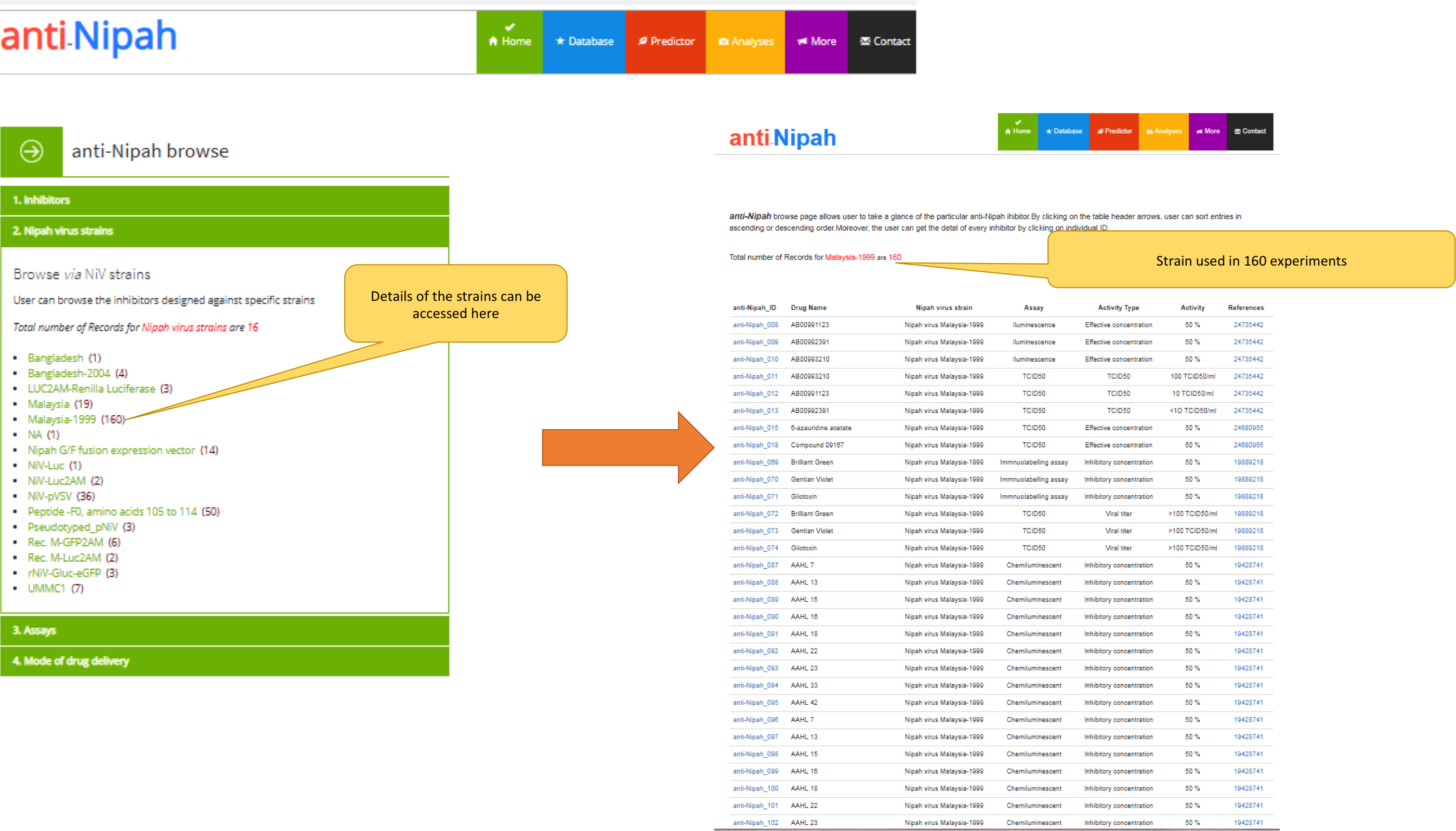
a) The "anti-Nipah" browse page allows the user to explore the anti-nipah molecules through drug name, NiV strain, assay and mode of drug delivery. The user can get the detail of each molecule by clicking on the entry id as shown in the image below.
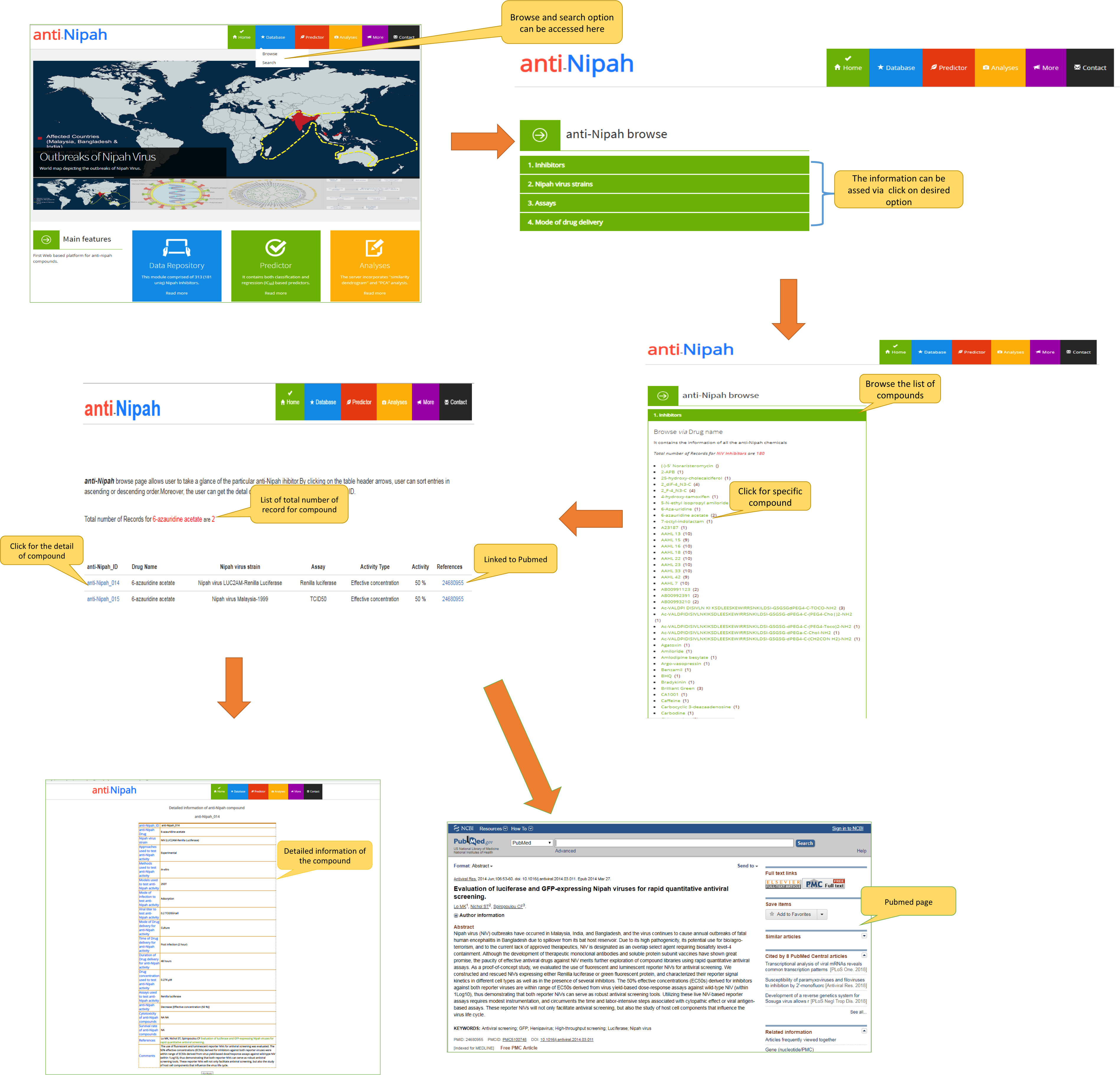
b) The page showing the browse option of the "anti-Nipah" via NiV strains
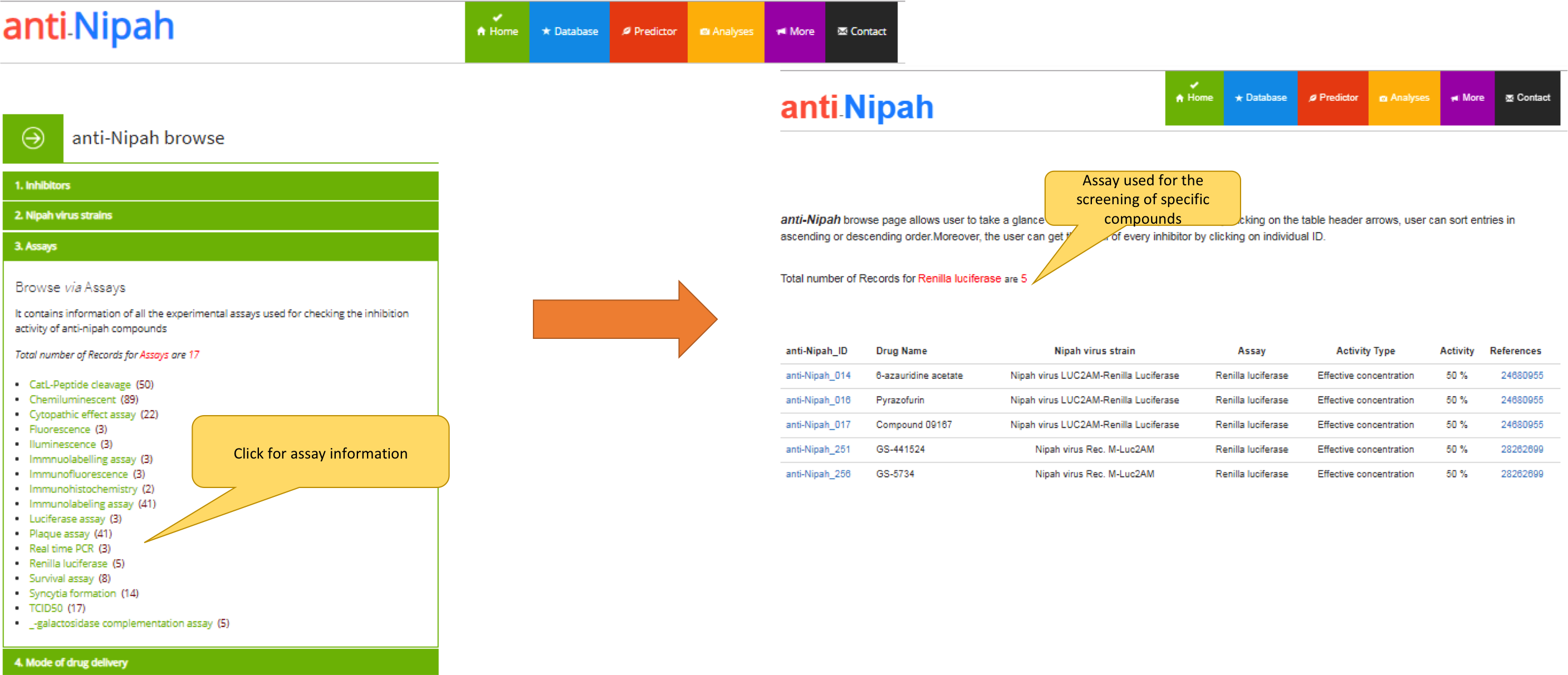
c) The page showing the browse option of the "anti-Nipah" via Assays
3. Search page
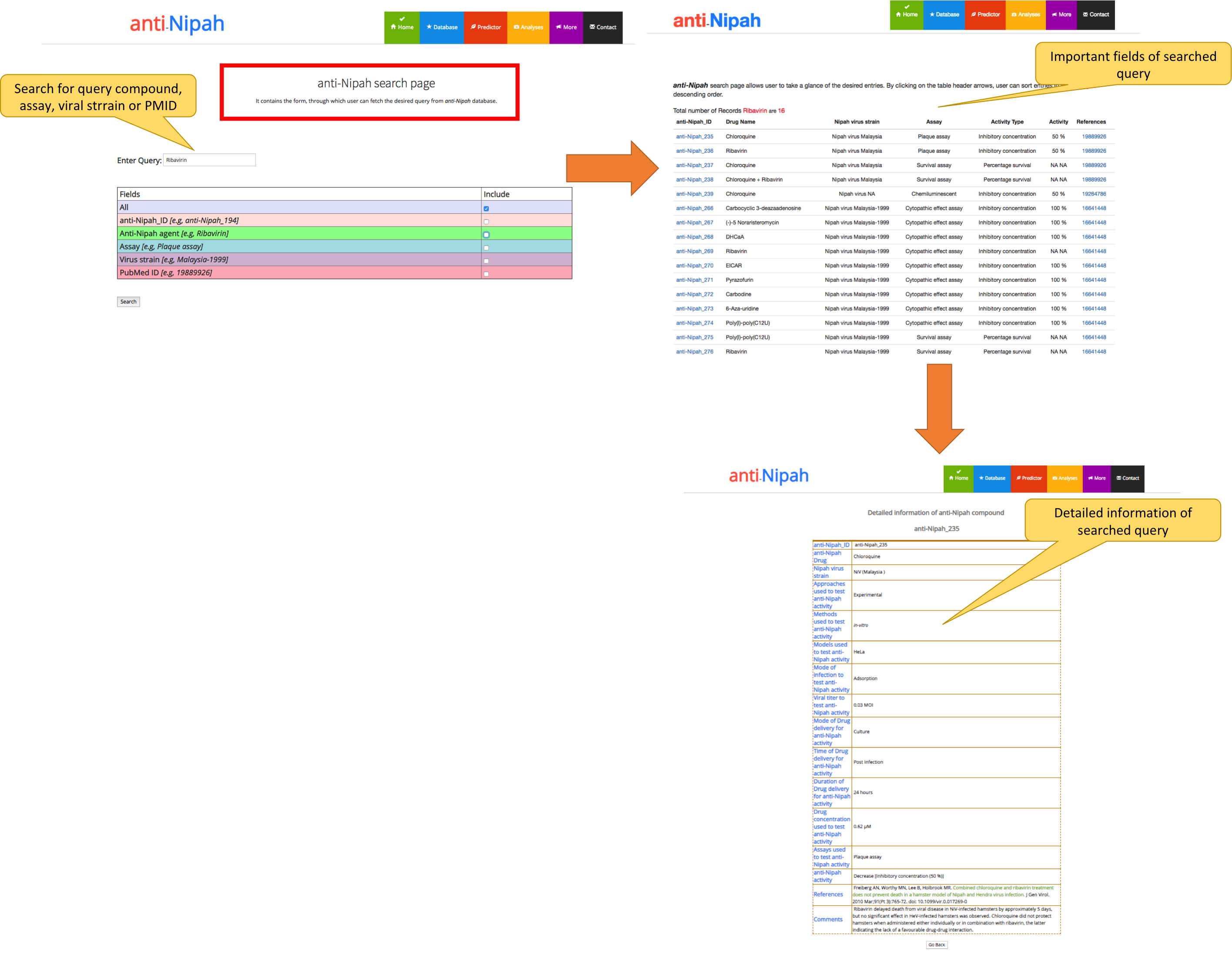
This section of the webserver can be useful to the user to search the desired query from the database. The user can search the query in all the fields or in specific fields like IDs, anti-Nipah agents, Assays, NiV strain, and PubMed ID
4. Predictor page
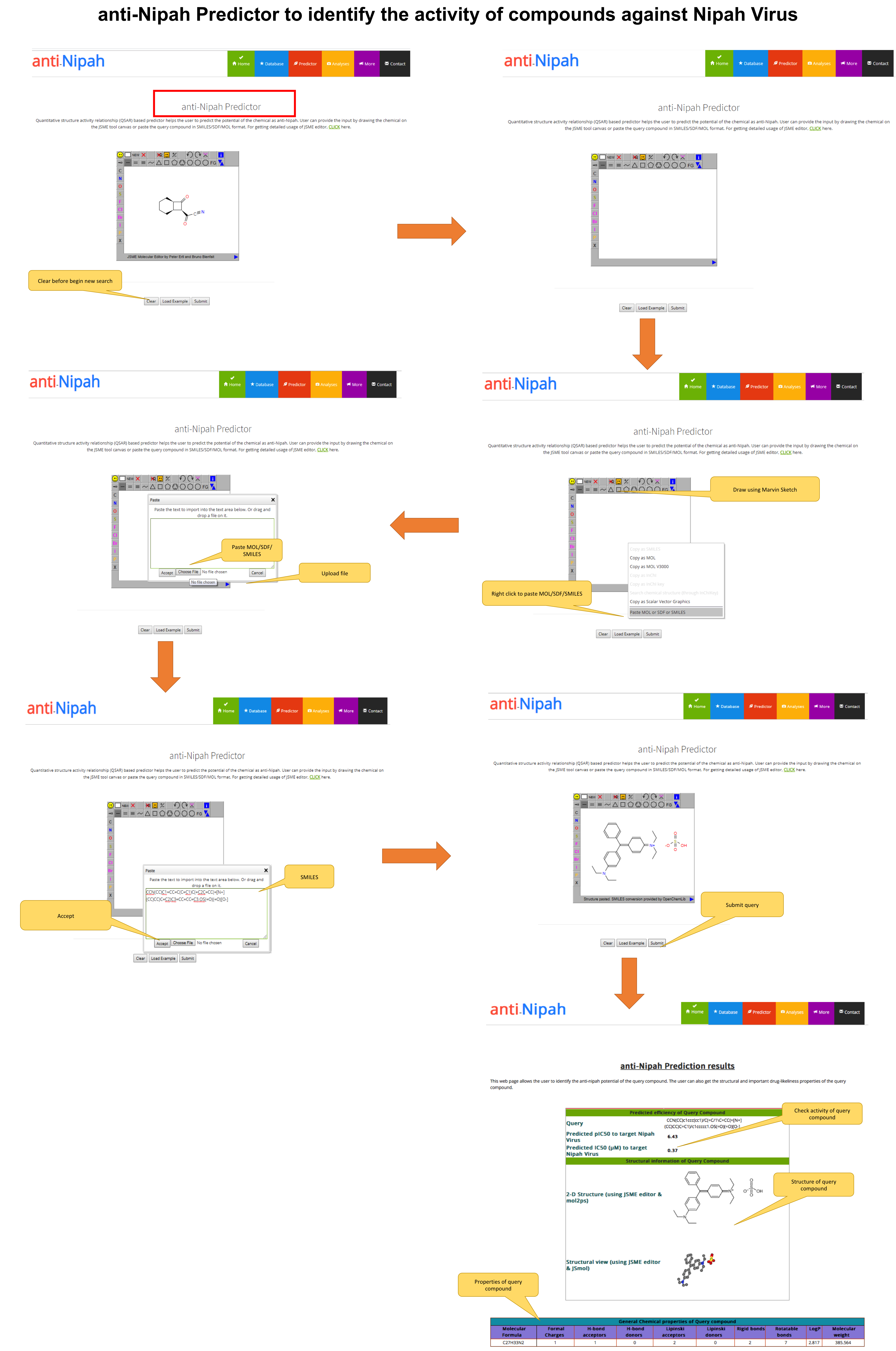
If the user has any chemical and want to predict its anti-Nipah activity. It can be accomplished with the help of this web page. The User can draw the query molecule or paste the SMILES/SDF/MOL file and submit it. Further the query molecule will be processed and classified on the developed model (QSAR), for checking its anti-Nipah activity.
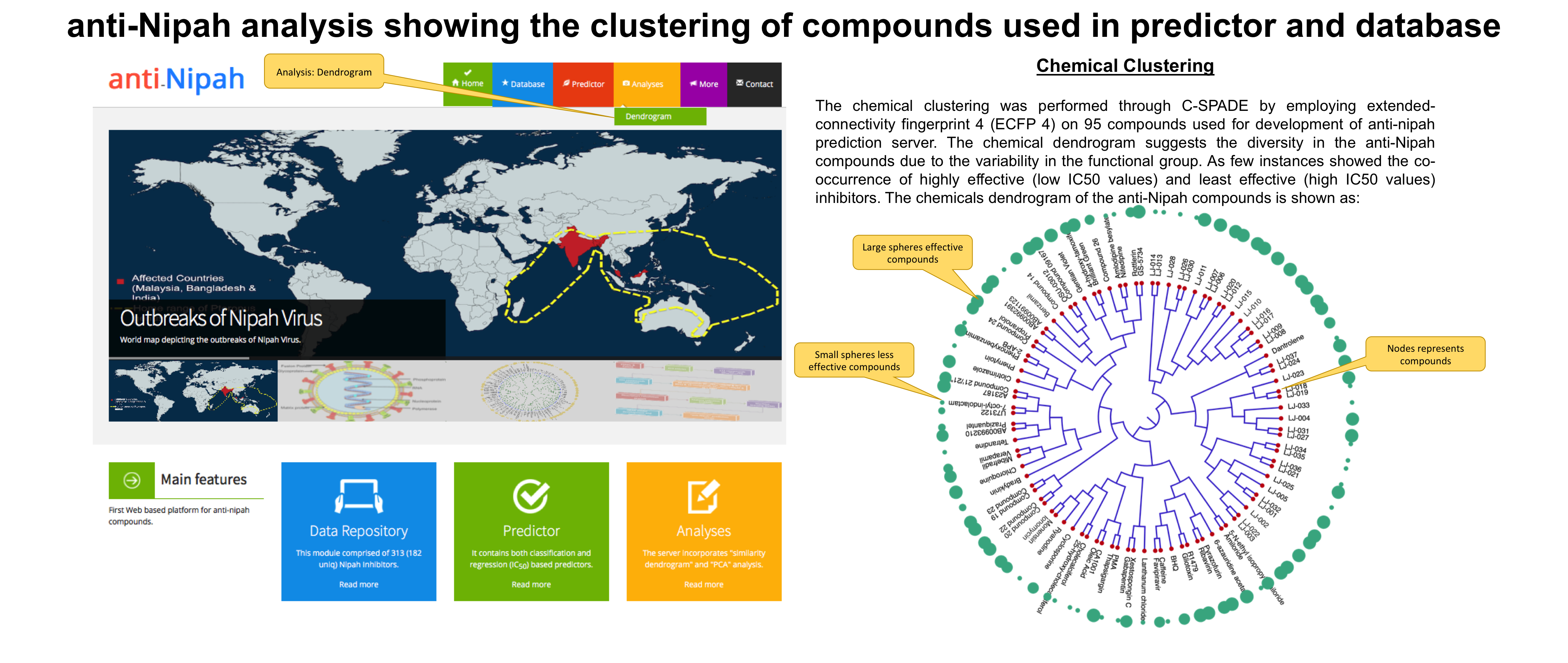
5. Analysis page
The data visualization module of the "anti-Nipah" is very useful for the users. It shows the chemical clustering via:
Extended connectivity fingerprint 4 through C-SPADE software